
Concept explainers
Bacterial genomes frequently contain groups of genes organized into operons. What is the biological advantage of operons to bacteria? Identify the regulatory components you would expect to find in an operon. How are the expressed genes of an operon usually arranged?
To review:
The genome of bacteria contains operons that consist of a group of genes. Describe the advantage of these operons to bacteria. Explain the expected regulatory components present in an operon. Also illustrate the arrangement of expressed genes of an operon?
Introduction:
The first operon was discovered in E. coli - “lac operon”- by a group of scientists- Francois Jacob, Andre Michel Lwoff and Jacques Monod in
Explanation of Solution
The gene expression in prokaryotes is completely dependent on the availability of the nutrients in a cell. To save energy and upsurge efficiency, bacteria have operons. The stretch of DNA that regulates the activity of structural genes and a group of genes is referred to as operons. They are regulated by positive and negative gene regulation. A single operon will give expressions of many gene products that save the energy of bacteria and increase the efficiency by gaining large amount of functional proteins. As bacteria have coupled transcription and translation, as soon as the group of genes on an operon is transcribed, they are converted to proteins. The genes are located one after another which allows protein synthesis to be controlled under a single promoter. To control the expression, the operon is switched on or off, according to the need of a cell.
The operon has regulatory components- Promoter, repressor, and activators/inducer.
The expressed genes are arranged after the regulatory region of the gene in a fashion one after another.
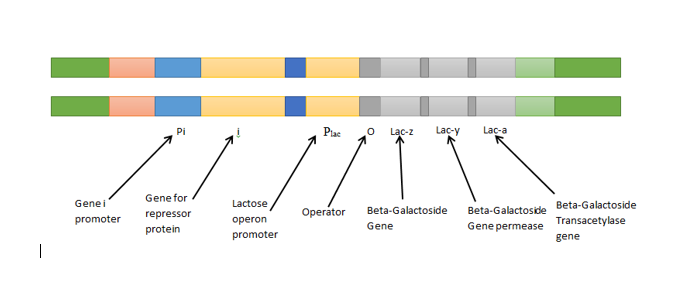
Operons in bacteria are described and the subsequent facts related to it are explained.
Want to see more full solutions like this?
Chapter 14 Solutions
Genetic Analysis: An Integrated Approach (2nd Edition)
- If a researcher moves the promoter for the lac operon to the region between the beta galactosidase (lacZ) gene and the permease (lacY) gene, which of the following results would you expect? A) The three genes of the lac operon will be expressed normally. B) The repressor will not be able to bind to the operon. C) The operon will still transcribe the lacZ and lacY genes, but the mRNA will not be translated. D) LacZ will not be transcribed and Beta galactosidase will not be produced.arrow_forwardWhat is the phenotype of an E. coli strain with a mutation in the lac operator that allows the lac repressor to irreversibly bind the operator? Assume glucose is absent. 1S A) O Transcription of the operon is very high whether lactose is present or not. B) O Transcription of the operon is very low whether lactose is present or not. C) O Transcription of the operon is high in the presence of lactose, and low in its absence. D) O Transcription of the operon is low in the presence of lactose, and high in its absence.arrow_forwardWhat are the effects of the following conditions on Lac operon of bacteria? Do not forget to mention about the role of repressor, activator, RNA polymerase in each case! A) Glucose is absent and lactose is present B) Glucose is present and lactose is present C) Glucose is present and lactose is absentarrow_forward
- Compare the types of bacterial genes associated with inducible operons, those associated with repressible operons, and those that are constitutive. Predict the category into which each of the following would most likely fit: (a) a gene that codes for RNA polymerase, (b) a gene that codes for an enzyme required to break down maltose, and (c) a gene that codes for an enzyme used in the synthesis of adenine.arrow_forwardThe attenuation mechanism that helps block expression of the tryptophan operon requires all of the following, except A) O MRNA transcript of the trp leader sequence B) Oaribosome stalled at the stop codon of the trp leader transcript C) O formation of an attenuator loop D) O low intracellular levels of tryptophanarrow_forwardYou are growing E. coli in a laboratory in order to study their operons. The growth media you are using contains lactose, no glucose and no tryptophan. Using your knowledge of operons and their regulation, Please explain so I can understand in the answer What effect does lactose have on the bacterial cell’s lac operon? What does the absence of glucose do to the bacterial cell? You now add tryptophan to the cell. What would happen to the bacterial cell and its trp operon? What kind of regulation does lactose provide to an operon’s repressor?arrow_forward
- The streptolysin S toxin made by S. pyogenes is encoded by a 9-gene operon, sagABCDEFGHI. Thinking about what a 3-line diagram would look like for this operon, answer the following questions. Write numeric answers only. For example, if your answer is 6 promoters, write only 6. 1) How many promoters control the expression of these genes? 2) How many locations does RNA Polymerase bind to get full expression of these genes? 3) How many ribosome binding sites are needed for full protein expression? 4) How many start codons will be needed for full protein expression? 5) How many mRNA strands will be produced with full operon expression? 6) How many proteins will be produced with full protein expression? 1arrow_forwardFigure 5 shows the lac operon structure in Escherichia coli. a) Name structures P, Q and R. b) Name substance S. c) What is the enzyme encoded by gene I. Give its function. d) What will happen if substance S is absent in the medium?arrow_forwardA lac operon containing one mutation was cloned into a plasmid, which was introduced by transformation into a bacterium containing a wild-type lac operon. The three genes of the chromosomal operon were rendered noninducible in the presence of the plasmid. (a) What kind of mutation in the plasmid operon could have this effect? (b) Suppose the result of transformation was to cause the three plasmid lac genes to be expressed constitutively, at a high level. What type of plasmid gene mutation could have this result?arrow_forward
- You have isolated different mutants (reg1 and reg2) causing constitutive expression of the emu operon (which has genes emu1 and emu2). One mutant contains a defect in a DNA-binding site, and the other has a loss-of-function defect in the gene encoding a protein that binds to the site Say you don’t know which mutant has a defect in the site and which one has a mutation in the binding protein. To figure it out, you construct the two partial diploid strains (i and ii below), and you then assay the levels of the Emu1 and Emu2 proteins in these two strains. F’ (reg1- reg2+ emu1- emu2+) / reg1+ reg2+ emu1+ emu2- F’ (reg1+ reg2- emu1- emu2+) / reg1+ reg2+ emu1+ emu2- What proteins do you predict will be expressed for strains i and ii if reg2 encodes the regulatory protein and reg1 is the regulatory site?arrow_forwardWhat is lac operon? Draw and/or identify the status of the lac operon in a given set of environmental conditions and/or cell mutations.arrow_forwardTwo closely linked open reading frames (ORFs) have been identified on the E. coli chromosome and they are predicted to encode genes sugX sugY which is involved in the catabolism (utilization) of a non-glucose sugar. In the figure below, the top line (with the big arrow) is the coding strand, and the bottom line is the non-coding (template) strand of the operon. Since the RNA sequence would be the “same” as the coding strand, please label (show the location of), in relation to the ORFs, the promoter(s) and 4 other genetic elements in this region that may be involved in the transcription and translation of sugX and sugY as well as the regulation of their expression. (Note: genetic elements are the regions of DNA or RNA.) If you are not comfortable with the coding strand of the DNA, feel free to draw the mRNA strand underneath and label the relevant features on the RNA strand, but the RNA strand must be aligned with the DNA strands in position and scale. Leader(s)/antileader(s) and…arrow_forward
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education





