
Concept explainers
Alkaptonuria is a human autosomal recessive disorder caused by mutation of the HAO gene that encodes the enzyme homogentisic acid oxidase. Restriction mapping
of the HAO gene region reveals four BamHI restriction sites
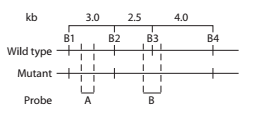
DNA samples taken from a mother (M), father (F), and two children
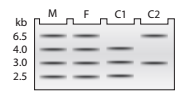
a. Using A to represent the
b. In a separate figure, draw the autoradiograph patterns for all the genotypes that could be found in children of this couple.
c. Explain how the DNA sequence change results in a
Want to see the full answer?
Check out a sample textbook solution
Chapter 12 Solutions
Genetic Analysis: An Integrated Approach (2nd Edition)
- This question has multiple parts (A, B & C), and under the subject of Nutrition. Thank you!arrow_forwardCalculate the CFU/ml of a urine sample if 138 E. coli colonies were counted on a Nutrient Agar Plate when0.5 mls were plated on the NA plate from a 10-9 dilution tube. You must highlight and express your answerin scientific notatioarrow_forwardDon't copy off the other answer if there is anyarrow_forward
- Use the following information to answer the question(s) below. Martin Wikelski and L. Michael Romero (Body size, performance and fitness in Galápagos marine iguanas, Integrative and Comparative Biology 43 [2003]:376-86) measured the snout-to-vent (anus) length of Galápagos marine iguanas and observed the percent survival of different-sized animals, all of the same age. The graph shows the log snout-vent length (SVL, a measure of overall body size) plotted against the percent survival of these different size classes for males and females. Survival (%) 100- 80- 60- 40- 20- 0+ 1.9 T 2 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Log SVL (mm) 19) Examine the figure above. What type of selection for body size appears to be occurring in these marine iguanas? A) directional selection B) stabilizing selection C) disruptive selection D) You cannot determine the type of selection from the above information. 3arrow_forward24) Use the following information to answer the question below. Researchers studying a small milkweed population note that some plants produce a toxin and other plants do not. They identify the gene responsible for toxin production. The dominant allele (T) codes for an enzyme that makes the toxin, and the recessive allele (t) codes for a nonfunctional enzyme that cannot produce the toxin. Heterozygotes produce an intermediate amount of toxin. The genotypes of all individuals in the population are determined (see table) and used to determine the actual allele frequencies in the population. TT 0.49 Tt 0.42 tt 0.09 Refer to the table above. Is this population in Hardy-Weinberg equilibrium? A) Yes. C) No; there are more homozygotes than expected. B) No; there are more heterozygotes than expected. D) It is impossible to tell.arrow_forward30) A B CDEFG Refer to the accompanying figure. Which of the following forms a monophyletic group? A) A, B, C, and D B) C and D C) D, E, and F D) E, F, and Garrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax





