
Concept explainers
(a)
Interpretation:
Lewis structure, VSEPR formula, bond angle, and molecular shape for
Concept Introduction:
Valence Shell Electron Pair Repulsion model predicts shape by inclusion of bond angles and most distant arrangement of atoms that leads to minimum repulsion. For the molecules that have no lone pairs around the central atom the bonded-atom unshared -pair arrangement is decided by the table as follows:
In order to determine the shape the steps to be followed are indicated as follows:
- 1. Lewis structure of molecule should be written.
- 2. The type electron arrangement around the central atom should be identified around the central atom. This essentially refers to determination of bond pairs and unshared or lone pairs around central atoms.
- 3. Then bonded-atom unshared -pair arrangement that can maximize the distance of electron pairs about central atom determines the shape.
For molecules that have lone pairs around central atom, lone pairs influence shape, because there are no atoms at the positions occupied by these lone pairs. The key rule that governs the molecular shape, in this case, is the extent of lone –lone pair repulsions are far greater than lone bond pair or bond pair-bond pair repulsions. The table that summarized the molecular shapes possible for various combinations of bonded and lone pairs are given as follows:
(a)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on each atom in
The skeleton structure in
These 12 electron pairs are allotted as lone pairs to satisfy respective octets. Hence, the Lewis structure in
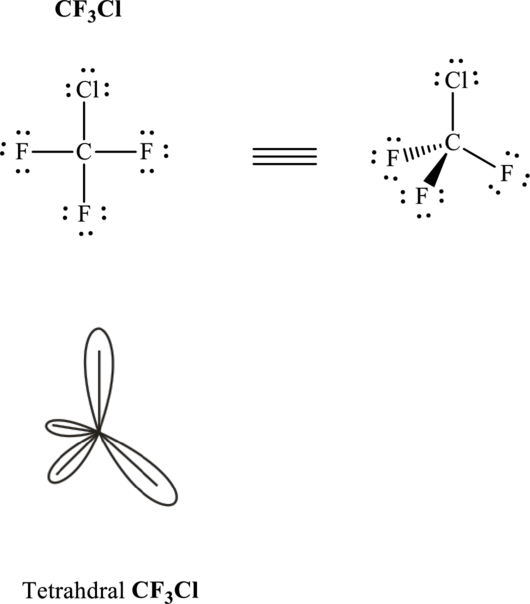
It is evident that in
If central atom is represented by A, and other attached bond pairs by X, then for any tetrahedral species with no lone pairs the VSEPR formula is predicted as
(b)
Interpretation:
Lewis structure, VSEPR formula, bond angle, and molecular shape for
Concept Introduction:
Refer to part (a).
(b)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on atom in
The skeleton structure in
These 13 electron pairs are allotted as lone pairs to satisfy respective octets. Hence, the Lewis structure in
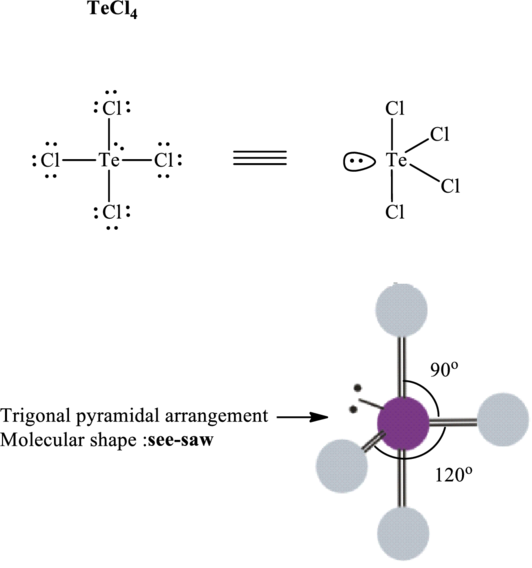
It is evident that
One lone pair is localized on equatorial positions so as to minimize lone pair–bond pair repulsions in accordance with VSPER model. This leads see-saw shape for
If lone pairs are represented by E, central atom with A and other attached bon pairs by X, then for any see-saw species the VSEPR formula is predicted to be
(c)
Interpretation:
Lewis structure, VSEPR formula, bond angle and molecular shape for
Concept Introduction:
Refer to part (a).
(c)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on atom in
The skeleton structure in
These 9 electron pairs are allotted as lone pairs or as multiple bonds to oxygen so as to satisfy octets. Hence, the Lewis structure in
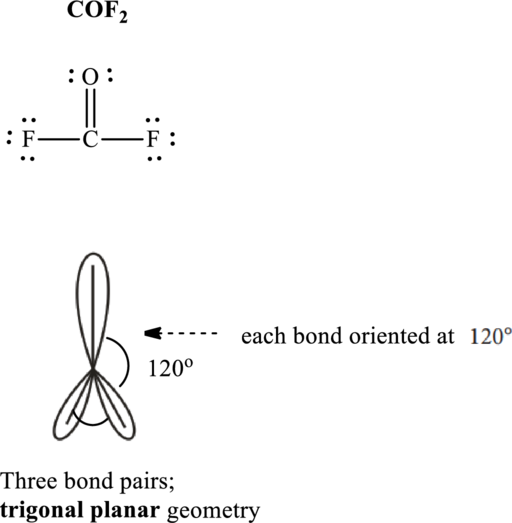
It is evident that in
If lone pairs are represented by E, central atom with A and other attached bond pairs by X, then for any trigonal planar species the VSEPR formula is predicted as
(d)
Interpretation:
Lewis structure, VSEPR formula, bond angle and molecular shape for
Concept Introduction:
Refer to part (a).
(d)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on atom in
Thus, Lewis structure in
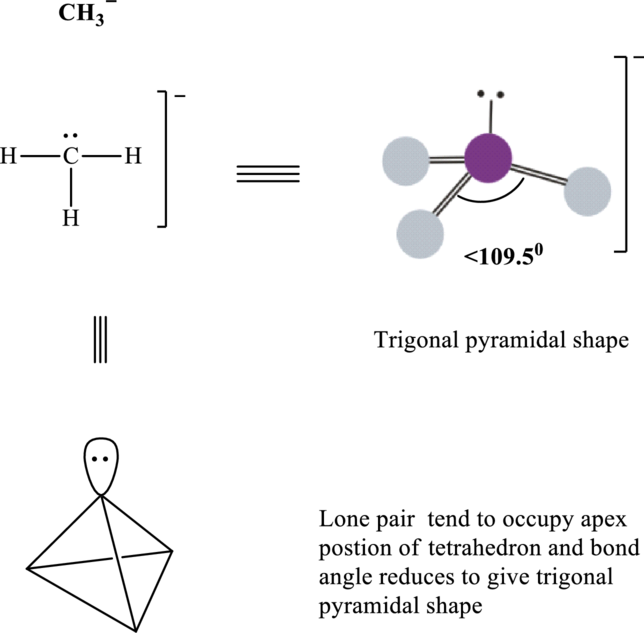
It is evident that in
Lone pair tend to be localized on apical position and so
If lone pairs are represented by E, central atom with A and other attached bond pairs by X, then for any trigonal pyramidal species the VSEPR formula is predicted as
Want to see more full solutions like this?
Chapter 2 Solutions
ACHIEVE/CHEMICAL PRINCIPLES ACCESS 2TERM
- Identify and provide a concise explanation of a specific analytical instrument capable of detecting and quantifying trace compounds in food samples. Emphasise the instrumental capabilities relevant to trace compound analysis in the nominated food. Include the specific application name (eg: identification and quantification of mercury in salmon), outline a brief description of sample preparation procedures, and provide a summary of the obtained results from the analytical process.arrow_forwardIdentify and provide an explanation of what 'Seperation Science' is. Also describe its importance with the respect to the chemical analysis of food. Provide specific examples.arrow_forward5. Propose a Synthesis for the molecule below. You may use any starting materials containing 6 carbons or less (reagents that aren't incorporated into the final molecule such as PhзP do not count towards this total, and the starting material can have whatever non-carbon functional groups you want), and any of the reactions you have learned so far in organic chemistry I, II, and III. Your final answer should show each step separately, with intermediates and conditions clearly drawn. H3C CH3arrow_forward
- State the name and condensed formula of isooxazole obtained by reacting acetylacetone and hydroxylamine.arrow_forwardState the name and condensed formula of the isothiazole obtained by reacting acetylacetone and thiosemicarbazide.arrow_forwardProvide the semi-developed formula of isooxazole obtained by reacting acetylacetone and hydroxylamine.arrow_forward
- Given a 1,3-dicarbonyl compound (R1-CO-CH2-CO-R2), indicate the formula of the compound obtaineda) if I add hydroxylamine (NH2OH) to give an isooxazole.b) if I add thiosemicarbazide (NH2-CO-NH-NH2) to give an isothiazole.arrow_forwardAn orange laser has a wavelength of 610 nm. What is the energy of this light?arrow_forwardThe molar absorptivity of a protein in water at 280 nm can be estimated within ~5-10% from its content of the amino acids tyrosine and tryptophan and from the number of disulfide linkages (R-S-S-R) between cysteine residues: Ε280 nm (M-1 cm-1) ≈ 5500 nTrp + 1490 nTyr + 125 nS-S where nTrp is the number of tryptophans, nTyr is the number of tyrosines, and nS-S is the number of disulfide linkages. The protein human serum transferrin has 678 amino acids including 8 tryptophans, 26 tyrosines, and 19 disulfide linkages. The molecular mass of the most dominant for is 79550. Predict the molar absorptivity of transferrin. Predict the absorbance of a solution that’s 1.000 g/L transferrin in a 1.000-cm-pathlength cuvet. Estimate the g/L of a transferrin solution with an absorbance of 1.50 at 280 nm.arrow_forward
- In GC, what order will the following molecules elute from the column? CH3OCH3, CH3CH2OH, C3H8, C4H10arrow_forwardBeer’s Law is A = εbc, where A is absorbance, ε is the molar absorptivity (which is specific to the compound and wavelength in the measurement), and c is concentration. The absorbance of a 2.31 × 10-5 M solution of a compound is 0.822 at a wavelength of 266 nm in a 1.00-cm cell. Calculate the molar absorptivity at 266 nm.arrow_forwardHow to calculate % of unknown solution using line of best fit y=0.1227x + 0.0292 (y=2.244)arrow_forward
 Chemistry: Principles and PracticeChemistryISBN:9780534420123Author:Daniel L. Reger, Scott R. Goode, David W. Ball, Edward MercerPublisher:Cengage Learning
Chemistry: Principles and PracticeChemistryISBN:9780534420123Author:Daniel L. Reger, Scott R. Goode, David W. Ball, Edward MercerPublisher:Cengage Learning Chemistry: The Molecular ScienceChemistryISBN:9781285199047Author:John W. Moore, Conrad L. StanitskiPublisher:Cengage Learning
Chemistry: The Molecular ScienceChemistryISBN:9781285199047Author:John W. Moore, Conrad L. StanitskiPublisher:Cengage Learning Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning
Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning


