
To review:
The type of inhibition by A and B inhibitor and the kind of affinity they possess for ES versus E.
Introduction:
Enzyme inhibitors are the molecules that are able to bind to enzyme and reduce the activity. Enzyme inhibitors work by several ways such as preventing the substrate from entering the active site of an enzyme or hinder in catalyzing the reaction. There are three types of enzyme inhibitors, namely, competitive, noncompetitive, and uncompetitive.
Explanation of Solution
The velocity of the reaction at different substrate concentrations in presence of A inhibitor and B inhibitor are given in the question. For calculating, the Vmax (maximum
| [S] in mM (millimolar) |
V | |
|
| 1.3 | 1.17 | 0.76 | 0.855 |
| 2.6 | 2.10 | 0.38 | 0.476 |
| 6.5 | 4.00 | 0.15 | 0.25 |
| 13.0 | 5.70 | 0.077 | 0.175 |
| 26.0 | 7.20 | 0.038 | 0.139 |
The following table shows the values of [S], V,
| [S] in mM (millimolar) |
V | |
|
| 1.3 | 0.62 | 0.76 | 1.613 |
| 2.6 | 1.42 | 0.38 | 0.704 |
| 6.5 | 2.65 | 0.15 | 0.377 |
| 13.0 | 3.12 | 0.077 | 0.321 |
| 26.0 | 3.58 | 0.038 | 0.28 |
Plotting the graph between
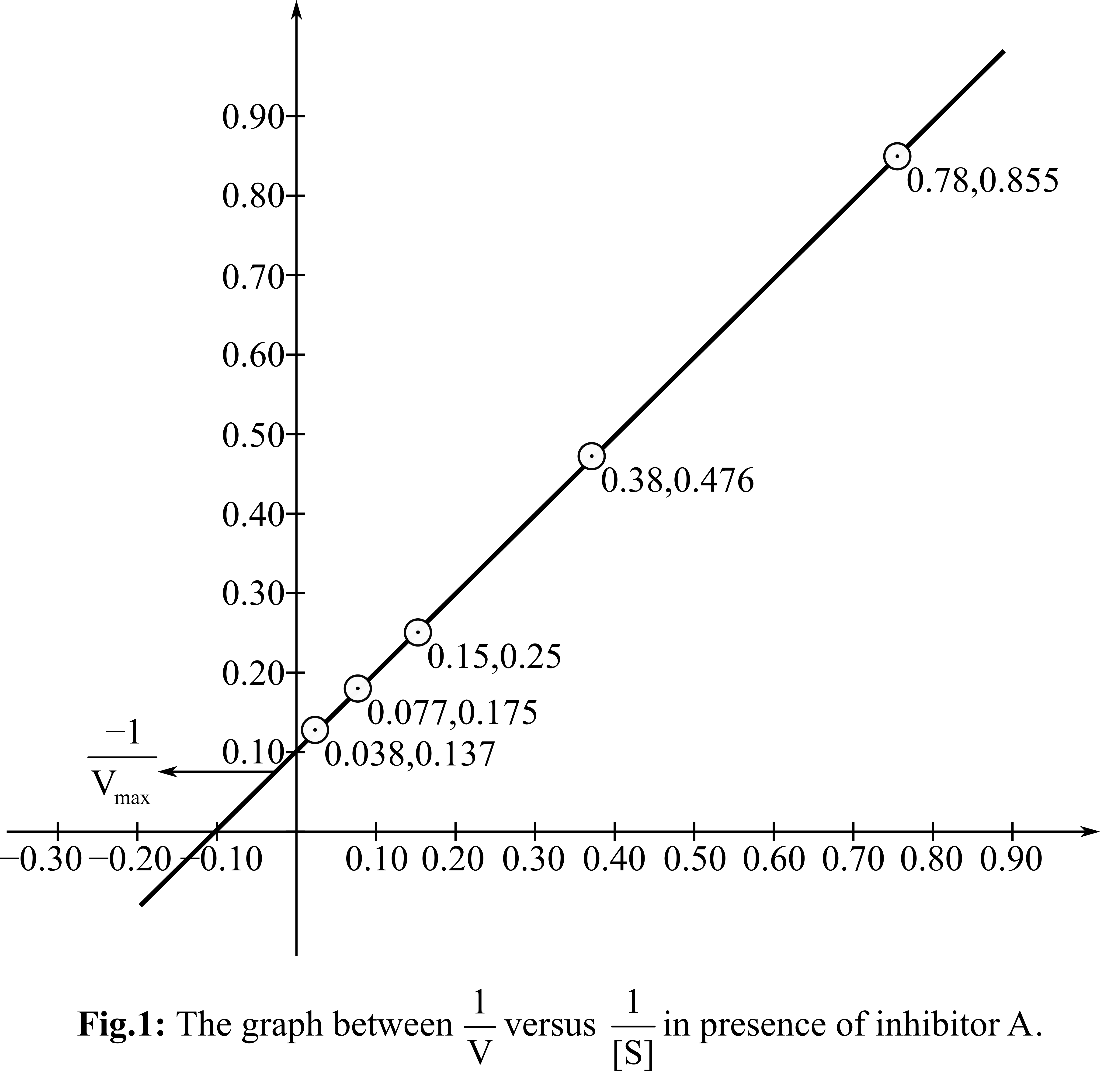
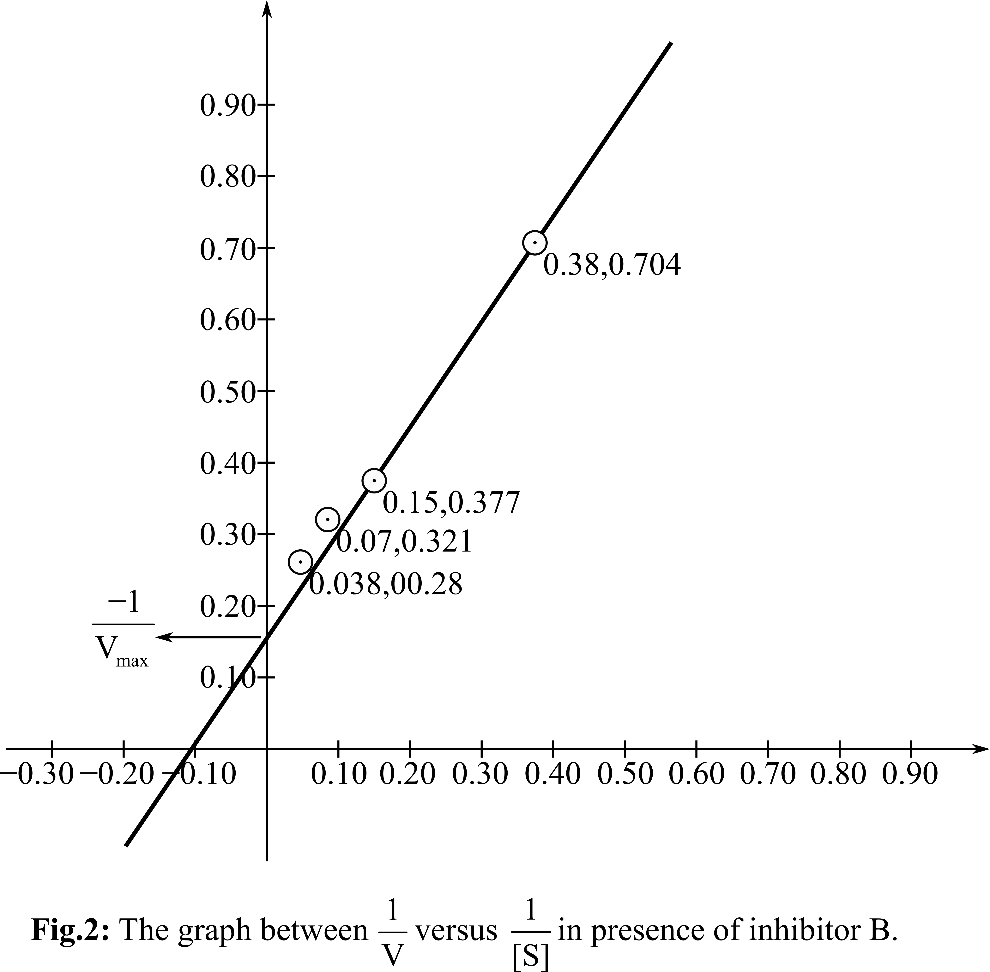
According, to the Michelis Menten equation, the value of y-intercept represents
From the graph, it can be observed that the value of y-intercept is 0.10 mM-1 sec. y-Intercept represents inverse of Vmax.
KM can be calculated by putting the value of Vmax in the equation Slope=KM/Vmax
So, the KM for the reaction is 9.8and the Vmax is 10 mM. sec-1. In presence of inhibitor A, KM is changed but Vmax is unchanged. So, it is a competitive inhibitor.
According, to the Michelis Menten equation, the value of y-intercept represents
From the graph, it can be observed that the value of y-intercept is 0.25 mM-1 sec. y-Intercept represents inverse of Vmax.
KM can be calculated by putting the value of Vmax in the equation Slope=KM/Vmax
So, the KM for the reaction is 5.68 and the Vmax is4 mM. sec-1. In presence of inhibitor B, KM andVmax both are changed. So, it is anuncompetitive inhibitor. A inhibitor is a competitive inhibitor so it can bind to free enzyme and has no affinity for ES. B inhibitor is an uncompetitive inhibitor, so it can bind to ES as well as E.
Therefore, it can be concluded that inhibitor A is competitive as Vmax remain unchanged whereas, inhibitor B is uncompetitive inhibitor. A can bind to free enzyme and B can bind to E as well as ES.
Want to see more full solutions like this?
Chapter 6 Solutions
Biochemistry, The Molecular Basis of Life, 6th Edition
- please draw it for me and tell me where i need to modify the structurearrow_forwardPlease help determine the standard curve for my Kinase Activity in Excel Spreadsheet. Link: https://mnscu-my.sharepoint.com/personal/vi2163ss_go_minnstate_edu/_layouts/15/Doc.aspx?sourcedoc=%7B958f5aee-aabd-45d7-9f7e-380002892ee0%7D&action=default&slrid=9b178ea1-b025-8000-6e3f-1cbfb0aaef90&originalPath=aHR0cHM6Ly9tbnNjdS1teS5zaGFyZXBvaW50LmNvbS86eDovZy9wZXJzb25hbC92aTIxNjNzc19nb19taW5uc3RhdGVfZWR1L0VlNWFqNVc5cXRkRm4zNDRBQUtKTHVBQldtcEtWSUdNVmtJMkoxQzl3dmtPVlE_cnRpbWU9eEE2X291ZHIzVWc&CID=e2126631-9922-4cc5-b5d3-54c7007a756f&_SRM=0:G:93 Determine the amount of VRK1 is present 1. Average the data and calculate the mean absorbance for each concentration/dilution (Please over look for Corrections) 2. Blank Correction à Subtract 0 ug/mL blank absorbance from all readings (Please over look for Corrections) 3. Plot the Standard Curve (Please over look for Corrections) 4. Convert VRK1 concentration from ug/mL to g/L 5. Use the molar mass of VRK1 to convert to M and uM…arrow_forwardMacmillan Learning Cholesterol synthesis begins with the formation of mevalonate from acetyl CoA. This process activates mevalonate and converts it to isopentenyl pyrophosphate. Identify the atoms in mevalonate and isopentenyl pyrophosphate that will be labeled from acetyl CoA labeled with 14C in the carbonyl carbon. Place 14C atoms and C atoms to denote which carbon atoms are labeled and which are not labeled. H₂C COA 14C-labeled acetyl-CoA HHH [c] H H OH 014C - OH H HH H Mevalonate CH3 H H 14C H Η H H Incorrect Answer of o -P-O-P-0- Isopentenyl pyrophosphate с Answer Bank 14Carrow_forward
- Draw the reaction between sphingosine and arachidonic acid. Draw out the full structures.arrow_forwardDraw both cis and trans oleic acid. Explain why cis-oleic acid has a melting point of 13.4°C and trans-oleic acid has a melting point of 44.5°C.arrow_forwardDraw the full structure of the mixed triacylglycerol formed by the reaction of glycerol and the fatty acids arachidic, lauric and trans-palmitoleic. Draw the line structure.arrow_forward
- Draw out the structure for lycopene and label each isoprene unit. "Where is lycopene found in nature and what health benefits does it provide?arrow_forwardWhat does it mean to be an essential fatty acid? What are the essential fatty acids?arrow_forwardCompare and contrast primary and secondary active transport mechanisms in terms of energy utilisation and efficiency. Provide examples of each and discuss their physiological significance in maintaining ionic balance and nutrient uptake. Rubric Understanding the key concepts (clearly and accurately explains primary and secondary active transport mechanisms, showing a deep understanding of their roles) Energy utilisation analysis ( thoroughly compares energy utilisation in primary and secondary transport with specific and relevant examples Efficiency discussion Use of examples (provides relevant and accurate examples (e.g sodium potassium pump, SGLT1) with clear links to physiological significance. Clarity and structure (presents ideas logically and cohesively with clear organisation and smooth transition between sections)arrow_forward
- 9. Which one of the compounds below is the major organic product obtained from the following reaction sequence, starting with ethyl acetoacetate? 요요. 1. NaOCH2CH3 CH3CH2OH 1. NaOH, H₂O 2. H3O+ 3. A OCH2CH3 2. ethyl acetoacetate ii A 3. H3O+ OH B C D Earrow_forward7. Only one of the following ketones cannot be made via an acetoacetic ester synthesis. Which one is it? Ph کہ A B C D Earrow_forward2. Which one is the major organic product obtained from the following reaction sequence? HO A OH 1. NaOEt, EtOH 1. LiAlH4 EtO OEt 2. H3O+ 2. H3O+ OH B OH OH C -OH HO -OH OH D E .CO₂Etarrow_forward
 BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Human Physiology: From Cells to Systems (MindTap ...BiologyISBN:9781285866932Author:Lauralee SherwoodPublisher:Cengage Learning
Human Physiology: From Cells to Systems (MindTap ...BiologyISBN:9781285866932Author:Lauralee SherwoodPublisher:Cengage Learning Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage Learning
Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage Learning Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning





