
Concept explainers
Feather color in parakeets is produced by the blending of pigments produced from two biosynthetic pathways shown below. Four independently assorting genes (A, B, C, and D) produce enzymes that catalyze separate steps of the pathways. For the questions below, use an uppercase letter to indicate a dominant allele producing full enzymatic activity and a lowercase letter to indicate a recessive allele producing no functional enzyme. Feather colors produced by mixing pigments are green (yellow + blue) and purple (red + blue). Red, yellow, and blue feathers result from production of one colored pigment, and white results from absence of pigment production.
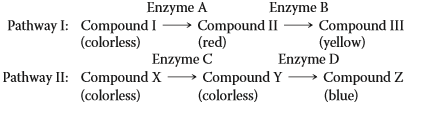
a. What is the genotype of a pure-breeding purple parakeet strain?
b. What is the genotype of a pure-breeding yellow strain of parakeet?
c. If a pure-breeding blue strain of parakeet (aa BB CC DD) is crossed to one that is pure-breeding purple, predict the genotype(s) and phenotype(s) of the
d. If
Learn your wayIncludes step-by-step video

Chapter 4 Solutions
Genetic Analysis: An Integrated Approach (3rd Edition)
Additional Science Textbook Solutions
Chemistry: Structure and Properties (2nd Edition)
Microbiology with Diseases by Body System (5th Edition)
Chemistry: The Central Science (14th Edition)
Human Biology: Concepts and Current Issues (8th Edition)
Chemistry: An Introduction to General, Organic, and Biological Chemistry (13th Edition)
Biology: Life on Earth with Physiology (11th Edition)
- calculate the questions showing the solution including variables,unit and equations all the questiosn below using the data a) B1, b) B2, c) hybrid rate constant (1) d) hybrid rate constant (2) e) t1/2,dist f) t1/2,elim g) k10 h) k12 i) k21 j) initial concentration (C0) k) central compartment volume (V1) l) steady-state volume (Vss) m) clearance (CL) AUC (0→10 min) using trapezoidal rule n) AUC (20→30 min) using trapezoidal rule o) AUCtail (AUC360→∞) p) total AUC (using short cut method) q) volume from AUC (VAUC)arrow_forwardQUESTION 8 For the following pedigree, assume that the mode of inheritance is X-linked recessive, and that the trait has full penetrance and expressivity and occurs at a very low frequency in the hum population. Using XA for the dominant allele and Xa for the recessive allele, assign genotypes for the following individuals (if it is not possible to figure out the second allele of a genotype, that with an underscore): 2 m 1 2 1 2 4 5 6 7 8 9 IV 1 2 3 5 6 7 8 CO 9 10 12 13 V 1, 2 3 4 5 6 7 8 9 10 11 12 13 a. Il-1: b. 11-2: c. III-3: d. III-4: e. If individuals IV-11 and IV-12 have another child, what is the probability that they will have a boy with the disorder?arrow_forwardAnswerrarrow_forward
- please,show workings ahd solutions calculate the questions showing the solution including variables,unit and equations all the questiosn below using the data a) B1, b) B2, c) hybrid rate constant (1) d) hybrid rate constant (2) e) t1/2,dist f) t1/2,elim g) k10 h) k12 i) k21 j) initial concentration (C0) k) central compartment volume (V1) l) steady-state volume (Vss) m) clearance (CL) AUC (0→10 min) using trapezoidal rule n) AUC (20→30 min) using trapezoidal rule o) AUCtail (AUC360→∞) p) total AUC (using short cut method) q) volume from AUC (VAUC)arrow_forwardQuestion about consensus and the relationship to transcriptional activityarrow_forwardQuestion about archaea and eukaryotic general transcription homologsarrow_forward
- Question about general transcription factors and their relationship to polymerasesarrow_forwardIdentify the indicated structure?arrow_forwardrewrite: Problem 1 (Mental Health): The survivor victim is dealing with acute stress and symptoms of a post-traumatic stress disorder (PTSD) due to their traumatic experience during the January 2025 wildfire. Goal 1: To alleviate the client's overall level, frequency, and intensity of anxiety and PTSD symptoms so that daily functioning remains unimpaired. Objective 1: The client will learn and regularly use at least two anxieties management techniques to reduce anxiety symptoms to less than three episodes per week. Intervention 1: The therapist will provide psychoeducation about anxiety and PTSD, including their symptoms and triggers. The therapist will also teach and assist the client in adopting relaxation techniques, such as deep breathing exercises and progressive muscle relaxation, to better manage anxiety and lessen PTSD symptoms.arrow_forward
- O Macmillan Learning You have 0.100 M solutions of acetic acid (pKa = 4.76) and sodium acetate. If you wanted to prepare 1.00 L of 0.100 M acetate buffer of pH 4.00, how many milliliters of acetic acid and sodium acetate would you add? acetic acid: mL sodium acetate: mLarrow_forwardHow does the cost of food affect the nutritional choices people make?arrow_forwardBiopharmaceutics and Pharmacokinetics:Two-Compartment Model Zero-Order Absorption Questions SHOW ALL WORK, including equation used, variables used and each step to your solution, report your regression lines and axes names (with units if appropriate) :Calculate a-q a) B1, b) B2, c) hybrid rate constant (1) d) hybrid rate constant (2) e) t1/2,dist f) t1/2,elim g) k10 h) k12 i) k21 j) initial concentration (C0) k) central compartment volume (V1) l) steady-state volume (Vss) m) clearance (CL) AUC (0→10 min) using trapezoidal rule n) AUC (20→30 min) using trapezoidal rule o) AUCtail (AUC360→∞) p) total AUC (using short cut method) q) volume from AUC (VAUC)arrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning





