
Concept explainers
(a)
Interpretation:
Lewis structure, VSEPR formula, bond angle, and molecular shape for
Concept Introduction:
Valence Shell Electron Pair Repulsion model predicts shape by inclusion of bond angles and most distant arrangement of atoms that leads to minimum repulsion. For the molecules that have no lone pairs around the central atom the bonded-atom unshared -pair arrangement is decided by the table as follows:
In order to determine the shape the steps to be followed are indicated as follows:
- 1. Lewis structure of molecule should be written.
- 2. The type electron arrangement around the central atom should be identified around the central atom. This essentially refers to determination of bond pairs and unshared or lone pairs around central atoms.
- 3. Then bonded-atom unshared -pair arrangement that can maximize the distance of electron pairs about central atom determines the shape.
For molecules that have lone pairs around central atom, lone pairs influence shape, because there are no atoms at the positions occupied by these lone pairs. The key rule that governs the molecular shape, in this case, is the extent of lone –lone pair repulsions are far greater than lone bond pair or bond pair-bond pair repulsions. The table that summarized the molecular shapes possible for various combinations of bonded and lone pairs are given as follows:
(a)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on each atom in
The skeleton structure in
These 12 electron pairs are allotted as lone pairs to satisfy respective octets. Hence, the Lewis structure in
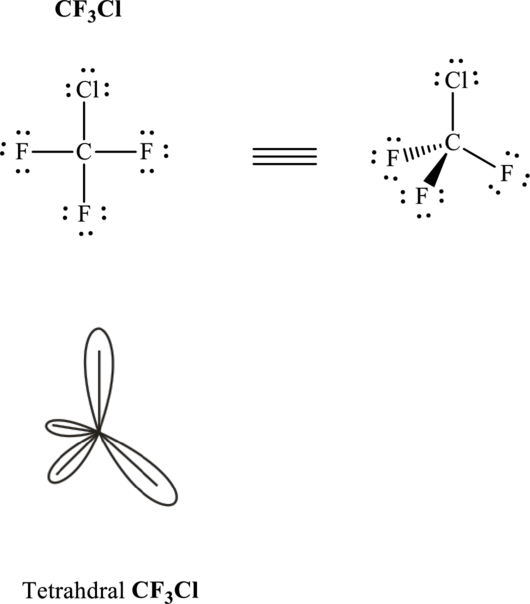
It is evident that in
If central atom is represented by A, and other attached bond pairs by X, then for any tetrahedral species with no lone pairs the VSEPR formula is predicted as
(b)
Interpretation:
Lewis structure, VSEPR formula, bond angle, and molecular shape for
Concept Introduction:
Refer to part (a).
(b)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on atom in
The skeleton structure in
These 13 electron pairs are allotted as lone pairs to satisfy respective octets. Hence, the Lewis structure in
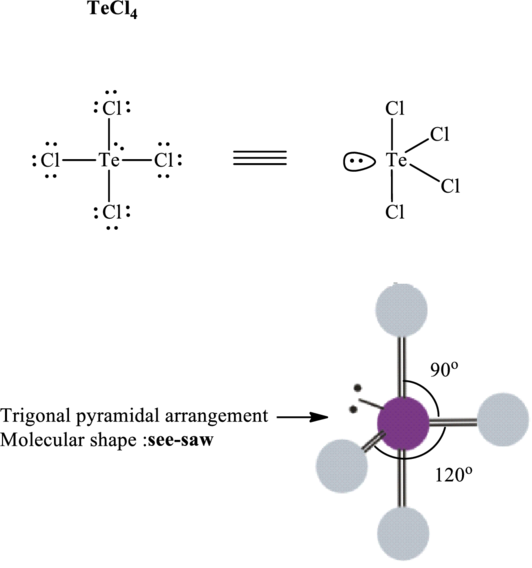
It is evident that
One lone pair is localized on equatorial positions so as to minimize lone pair–bond pair repulsions in accordance with VSPER model. This leads see-saw shape for
If lone pairs are represented by E, central atom with A and other attached bon pairs by X, then for any see-saw species the VSEPR formula is predicted to be
(c)
Interpretation:
Lewis structure, VSEPR formula, bond angle and molecular shape for
Concept Introduction:
Refer to part (a).
(c)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on atom in
The skeleton structure in
These 9 electron pairs are allotted as lone pairs or as multiple bonds to oxygen so as to satisfy octets. Hence, the Lewis structure in
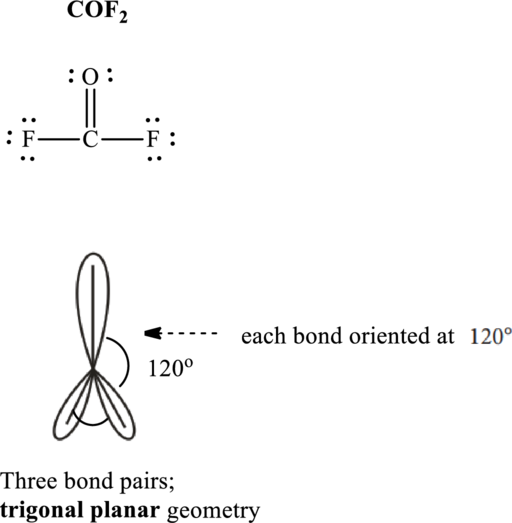
It is evident that in
If lone pairs are represented by E, central atom with A and other attached bond pairs by X, then for any trigonal planar species the VSEPR formula is predicted as
(d)
Interpretation:
Lewis structure, VSEPR formula, bond angle and molecular shape for
Concept Introduction:
Refer to part (a).
(d)
Answer to Problem 2E.15E
The shape for
Explanation of Solution
Total valence electrons are sum of the valence electrons on atom in
Thus, Lewis structure in
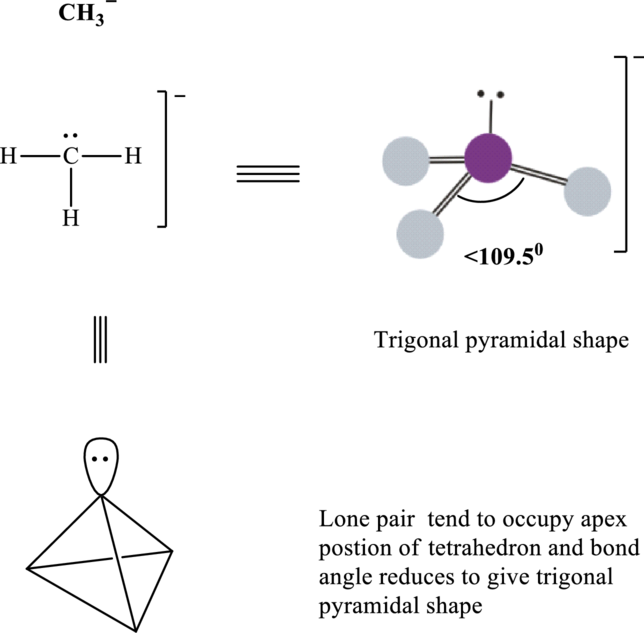
It is evident that in
Lone pair tend to be localized on apical position and so
If lone pairs are represented by E, central atom with A and other attached bond pairs by X, then for any trigonal pyramidal species the VSEPR formula is predicted as
Want to see more full solutions like this?
Chapter 2 Solutions
CHEM PRINCIPLES LL W/ACHIEVE ONE-SEM
- What are the major products of the following reaction? Please provide a detailed explanation and a drawing to show how the reaction proceeds.arrow_forwardWhat are the major products of the following organic reaction? Please include a detailed explanation as well as a drawing as to how the reaction proceeds.arrow_forwardPredict the organic product that forms in the reaction below: H + гон OH H+ H+ ☑ O Note: You may assume you have an excess of either reactant if the reaction requires more than one of those molecules to form the product. In the drawing area below, draw the skeletal ("line") structure of the missing organic product X. Explanation Check Click and drag to start drawing a structure. S 2025 McGraw Hill LLC. All Rights Reserved. Terms of Use | Privacy Centearrow_forward
- In the analysis of Mg content in a 25 mL sample, a titration volume of 5 mL was obtained using 0.01 M EDTA. Calculate the Mg content in the sample if the Ca content is 20 ppmarrow_forwardPredict the organic products that form in the reaction below: H. H+ + OH H+ Y Note: You may assume you have an excess of either reactant if the reaction requires more than one of those molecules to form the products. In the drawing area below, draw the skeletal ("line") structures of the missing organic products X and Y. You may draw the structures in any arrangement that you like, so long as they aren't touching. Explanation Check Click and drag to start drawing a structure. G X C © 2025 McGraw Hill LLC. All Rights Reserved. Terms of Use | Privacy Center | Access +arrow_forward111 Carbonyl Chem Choosing reagants for a Wittig reaction What would be the best choices for the missing reagents 1 and 3 in this synthesis? 1. PPh3 3 1 2 2. n-BuLi • Draw the missing reagents in the drawing area below. You can draw them in any arrangement you like. Do not draw the missing reagent 2. If you draw 1 correctly, we'll know what it is. • Note: if one of your reagents needs to contain a halogen, use bromine. Explanation Check Click and drag to start drawing a structure. × ©2025 McGraw Hill LLC. All Rights Reserved. Terms of Usearrow_forward
- A student proposes the transformation below in one step of an organic synthesis. There may be one or more reactants missing from the left-hand side, but there are no products missing from the right-hand side. There may also be catalysts, small inorganic reagents, and other important reaction conditions missing from the arrow. • Is the student's transformation possible? If not, check the box under the drawing area. . If the student's transformation is possible, then complete the reaction by adding any missing reactants to the left-hand side, and adding required catalysts, inorganic reagents, or other important reaction conditions above and below the arrow. • You do not need to balance the reaction, but be sure every important organic reactant or product is shown. + T X O O лет-ле HO OH HO OH This transformation can't be done in one step.arrow_forwardDetermine the structures of the missing organic molecules in the following reaction: X+H₂O H* H+ Y OH OH Note: Molecules that share the same letter have the exact same structure. In the drawing area below, draw the skeletal ("line") structures of the missing organic molecules X and Y. You may draw the structures in any arrangement that you like, so long as they aren't touching. Click and drag to start drawing a structure. X Sarrow_forwardPredict the major products of this organic reaction. If there aren't any products, because nothing will happen, check the box under the drawing area instead. No reaction. HO. O :☐ + G Na O.H Click and drag to start drawing a structure. XS xs H₂Oarrow_forward
- What are the angles a and b in the actual molecule of which this is a Lewis structure? H H C H- a -H b H Note for advanced students: give the ideal angles, and don't worry about small differences from the ideal groups may have slightly different sizes. a = b = 0 °arrow_forwardWhat are the angles a and b in the actual molecule of which this is a Lewis structure? :0: HCOH a Note for advanced students: give the ideal angles, and don't worry about small differences from the ideal that might be caused by the fact that different electron groups may have slightly different sizes. a = 0 b=0° Sarrow_forwardDetermine the structures of the missing organic molecules in the following reaction: + H₂O +H OH O OH +H OH X Note: Molecules that share the same letter have the exact same structure. In the drawing area below, draw the skeletal ("line") structure of the missing organic molecule X. Click and drag to start drawing a structure.arrow_forward
 Chemistry: Principles and PracticeChemistryISBN:9780534420123Author:Daniel L. Reger, Scott R. Goode, David W. Ball, Edward MercerPublisher:Cengage Learning
Chemistry: Principles and PracticeChemistryISBN:9780534420123Author:Daniel L. Reger, Scott R. Goode, David W. Ball, Edward MercerPublisher:Cengage Learning Chemistry: The Molecular ScienceChemistryISBN:9781285199047Author:John W. Moore, Conrad L. StanitskiPublisher:Cengage Learning
Chemistry: The Molecular ScienceChemistryISBN:9781285199047Author:John W. Moore, Conrad L. StanitskiPublisher:Cengage Learning Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning
Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning


