
Concept explainers
(a)
Interpretation:
The anti-codon on tRNA that should pair with UUG tRNA codon should be determined.
Concept Introduction:
The mRNA strand is arranged in a series of three (3) bases long units called codons. Each codons code for an amino acid. Corresponding to each unit of three (3) bases on the mRNA bo0und to ribosome, there is a complimentary set of three bases called anticodon, situated on the arm of tRNA molecule directly opposite to the arm that binds the amino acids. Marshall Nirenberg and Henrich Matthaei experimentally showed that by using a synthetic RNA chain consisting of only one type of base (uracil) a polypeptide chain containing only one type of amino acid, phenyl alanine is produced.
Thus, the base sequence UUU is the recognition site of phenylalanine. Similar model experiments with synthetic RNA molecules containing the other bases have yielded the entire RNA codes for all the 20 naturally occurring amino acids. Thus, the base sequence AUG-UUU-CAG-ACC-AAA in mRNA corresponds to the amino acid sequence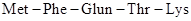 Several of the 64 combinations of these four (4) RNA bases (A, C, G, U) into groups of three to recognise only twenty (20) amino acids are redundant and do not code any amino acid signals the start and end of protein synthesis.
Several of the 64 combinations of these four (4) RNA bases (A, C, G, U) into groups of three to recognise only twenty (20) amino acids are redundant and do not code any amino acid signals the start and end of protein synthesis.
(b)
Interpretation:
The anti-codon on tRNA that should pair with CAC tRNA codon should be determined.
Concept Introduction:
The mRNA strand is arranged in a series of three (3) bases long units called codons. Each codons code for an amino acid. Corresponding to each unit of three (3) bases on the mRNA bo0und to ribosome, there is a complimentary set of three bases called anticodon, situated on the arm of tRNA molecule directly opposite to the arm that binds the amino acids. Marshall Nirenberg and Henrich Matthaei experimentally showed that by using a synthetic RNA chain consisting of only one type of base (uracil) a polypeptide chain containing only one type of amino acid, phenyl alanine is produced.
Thus, the base sequence UUU is the recognition site of phenylalanine. Similar model experiments with synthetic RNA molecules containing the other bases have yielded the entire RNA codes for all the 20 naturally occurring amino acids. Thus, the base sequence AUG-UUU-CAG-ACC-AAA in mRNA corresponds to the amino acid sequence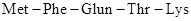 Several of the 64 combinations of these four (4) RNA bases (A, C, G, U) into groups of three to recognise only twenty (20) amino acids are redundant and do not code any amino acid signals the start and end of protein synthesis.
Several of the 64 combinations of these four (4) RNA bases (A, C, G, U) into groups of three to recognise only twenty (20) amino acids are redundant and do not code any amino acid signals the start and end of protein synthesis.
Want to see the full answer?
Check out a sample textbook solution
Chapter 16 Solutions
EBK CHEMISTRY FOR CHANGING TIMES
- 4. For the reactions below, draw the expected product. Be sure to indicate relevant stereochemistry or formal charges in the product structure. a) CI, H e b) H lux ligh Br 'Harrow_forwardArrange the solutions in order of increasing acidity. (Note that K (HF) = 6.8 x 10 and K (NH3) = 1.8 × 10-5) Rank solutions from least acidity to greatest acidity. To rank items as equivalent, overlap them. ▸ View Available Hint(s) Least acidity NH&F NaBr NaOH NH,Br NaCIO Reset Greatest acidityarrow_forward1. Consider the following molecular-level diagrams of a titration. O-HA molecule -Aion °° о ° (a) о (b) (c) (d) a. Which diagram best illustrates the microscopic representation for the EQUIVALENCE POINT in a titration of a weak acid (HA) with sodium. hydroxide? (e)arrow_forward
- Answers to the remaining 6 questions will be hand-drawn on paper and submitted as a single file upload below: Review of this week's reaction: H₂NCN (cyanamide) + CH3NHCH2COOH (sarcosine) + NaCl, NH4OH, H₂O ---> H₂NC(=NH)N(CH3)CH2COOH (creatine) Q7. Draw by hand the reaction of creatine synthesis listed above using line structures without showing the Cs and some of the Hs, but include the lone pairs of electrons wherever they apply. (4 pts) Q8. Considering the Zwitterion form of an amino acid, draw the Zwitterion form of Creatine. (2 pts) Q9. Explain with drawing why the C-N bond shown in creatine structure below can or cannot rotate. (3 pts) NH2(C=NH)-N(CH)CH2COOH This bond Q10. Draw two tautomers of creatine using line structures. (Note: this question is valid because problem Q9 is valid). (4 pts) Q11. Mechanism. After seeing and understanding the mechanism of creatine synthesis, students should be ready to understand the first half of one of the Grignard reactions presented in a past…arrow_forwardPropose a synthesis pathway for the following transformations. b) c) d)arrow_forwardThe rate coefficient of the gas-phase reaction 2 NO2 + O3 → N2O5 + O2 is 2.0x104 mol–1 dm3 s–1 at 300 K. Indicate whether the order of the reaction is 0, 1, or 2.arrow_forward
- 8. Draw all the resonance forms for each of the following molecules or ions, and indicate the major contributor in each case, or if they are equivalent. (4.5 pts) (a) PH2 سمةarrow_forward3. Assign absolute configuration (Rors) to each chirality center. a. H Nitz C. он b. 0 H-C. C H 7 C. ་-4 917-417 refs H 1つ ८ ડુ d. Но f. -2- 01 Ho -OH 2HNarrow_forwardHow many signals do you expect in the H NMR spectrum for this molecule? Br Br Write the answer below. Also, in each of the drawing areas below is a copy of the molecule, with Hs shown. In each copy, one of the H atoms is colored red. Highlight in red all other H atoms that would contribute to the same signal as the H already highlighted red. Note for advanced students: In this question, any multiplet is counted as one signal. Number of signals in the 'H NMR spectrum. For the molecule in the top drawing area, highlight in red any other H atoms that will contribute to the same signal as the H atom already highlighted red. If no other H atoms will contribute, check the box at right. No additional Hs to color in top molecule For the molecule in the bottom drawing area, highlight in red any other H atoms that will contribute to the same signal as the H atom already highlighted red. If no other H atoms will contribute, check the box at right. No additional Hs to color in bottom moleculearrow_forward
- In the drawing area below, draw the major products of this organic reaction: 1. NaOH ? 2. CH3Br If there are no major products, because nothing much will happen to the reactant under these reaction conditions, check the box under the drawing area instead. No reaction. Click and drag to start drawing a structure. ☐ : A คarrow_forwardPredict the major products of the following organic reaction: NC Δ ? Some important Notes: • Draw the major product, or products, of the reaction in the drawing area below. • If there aren't any products, because no reaction will take place, check the box below the drawing area instead. • Be sure to draw bonds carefully to show important geometric relationships between substituents. Note: if your answer contains a complicated ring structure, you must use one of the molecular fragment stamps (available in the menu at right) to enter the ring structure. You can add any substituents using the pencil tool in the usual way. Click and drag to start drawing a structure. Х аarrow_forwardPredict the major products of this organic reaction. Be sure you use dash and wedge bonds to show stereochemistry where it's important. + ☑ OH 1. TsCl, py .... 文 P 2. t-BuO K Click and drag to start drawing a structure.arrow_forward
 ChemistryChemistryISBN:9781305957404Author:Steven S. Zumdahl, Susan A. Zumdahl, Donald J. DeCostePublisher:Cengage Learning
ChemistryChemistryISBN:9781305957404Author:Steven S. Zumdahl, Susan A. Zumdahl, Donald J. DeCostePublisher:Cengage Learning ChemistryChemistryISBN:9781259911156Author:Raymond Chang Dr., Jason Overby ProfessorPublisher:McGraw-Hill Education
ChemistryChemistryISBN:9781259911156Author:Raymond Chang Dr., Jason Overby ProfessorPublisher:McGraw-Hill Education Principles of Instrumental AnalysisChemistryISBN:9781305577213Author:Douglas A. Skoog, F. James Holler, Stanley R. CrouchPublisher:Cengage Learning
Principles of Instrumental AnalysisChemistryISBN:9781305577213Author:Douglas A. Skoog, F. James Holler, Stanley R. CrouchPublisher:Cengage Learning Organic ChemistryChemistryISBN:9780078021558Author:Janice Gorzynski Smith Dr.Publisher:McGraw-Hill Education
Organic ChemistryChemistryISBN:9780078021558Author:Janice Gorzynski Smith Dr.Publisher:McGraw-Hill Education Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning
Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning Elementary Principles of Chemical Processes, Bind...ChemistryISBN:9781118431221Author:Richard M. Felder, Ronald W. Rousseau, Lisa G. BullardPublisher:WILEY
Elementary Principles of Chemical Processes, Bind...ChemistryISBN:9781118431221Author:Richard M. Felder, Ronald W. Rousseau, Lisa G. BullardPublisher:WILEY





