
EBK GENETICS: FROM GENES TO GENOMES
6th Edition
ISBN: 9781260041255
Author: HARTWELL
Publisher: MCGRAW HILL BOOK COMPANY
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 11, Problem 30P
Now consider a mating between consanguineous people involving a recessive genetic disease. The figures that follow show the genotypes of these two people at four SNP loci (1–4).
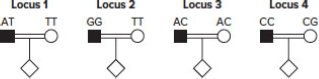
| a. | For which of these loci is it possible to obtain information about the linkage of the SNP to the disease gene? Explain your answer for each locus and describe any special conditions that may apply. |
| b. | For any of the loci for which the mating is potentially informative, how would you tell whether the child is the product of a recombinant or non recombinant gamete? (That is, how could you solve the phase problem?) Be as specific as possible. |
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Identify the indicated structure?
rewrite:
Problem 1 (Mental Health): The survivor victim is dealing with acute stress and symptoms of a
post-traumatic stress disorder (PTSD) due to their traumatic experience during the January 2025 wildfire.
Goal 1: To alleviate the client's overall level, frequency, and intensity of
anxiety and PTSD symptoms so that daily functioning remains
unimpaired.
Objective 1: The client will learn and regularly use at least two anxieties
management techniques to reduce anxiety symptoms to less than three episodes per week.
Intervention 1: The therapist will provide psychoeducation about anxiety and
PTSD, including their symptoms and triggers. The therapist will also teach and assist the client in adopting relaxation techniques, such as deep breathing exercises and progressive muscle relaxation, to better manage anxiety and lessen PTSD symptoms.
O Macmillan Learning
You have 0.100 M solutions of acetic acid (pKa = 4.76) and sodium acetate. If you wanted to prepare 1.00 L of 0.100 M acetate
buffer of pH 4.00, how many milliliters of acetic acid and sodium acetate would you add?
acetic acid:
mL
sodium acetate:
mL
Chapter 11 Solutions
EBK GENETICS: FROM GENES TO GENOMES
Ch. 11 - Choose the phrase from the right column that best...Ch. 11 - Would you characterize the pattern of inheritance...Ch. 11 - Would you be more likely to find single nucleotide...Ch. 11 - A recent estimate of the rate of base...Ch. 11 - If you examine Fig. 11.5 closely, you will note...Ch. 11 - Approximately 50 million SNPs have thus far been...Ch. 11 - Mutations at simple sequence repeat SSR loci occur...Ch. 11 - Humans and gorillas last shared a common ancestor...Ch. 11 - In 2015, an international team of scientists...Ch. 11 - Using PCR, you want to amplify an approximately 1...
Ch. 11 - Prob. 11PCh. 11 - The previous problem raises several interesting...Ch. 11 - You want to make a recombinant DNA in which a PCR...Ch. 11 - You sequence a PCR product amplified from a...Ch. 11 - Prob. 15PCh. 11 - The trinucleotide repeat region of the Huntington...Ch. 11 - Sperm samples were taken from two men just...Ch. 11 - Prob. 18PCh. 11 - a. It is possible to perform DNA fingerprinting...Ch. 11 - On July 17, 1918, Tsar Nicholas II; his wife the...Ch. 11 - The figure that follows shows DNA fingerprint...Ch. 11 - Microarrays were used to determine the genotypes...Ch. 11 - A partial sequence of the wild-type HbA allele is...Ch. 11 - a. In Fig. 11.17b, PCR is performed to amplify...Ch. 11 - The following figure shows a partial microarray...Ch. 11 - Scientists were surprised to discover recently...Ch. 11 - The microarray shown in Problem 25 analyzes...Ch. 11 - The figure that follows shows the pedigree of a...Ch. 11 - One of the difficulties faced by human geneticists...Ch. 11 - Now consider a mating between consanguineous...Ch. 11 - The pedigree shown in Fig. 11.22 was crucial to...Ch. 11 - You have identified a SNP marker that in one large...Ch. 11 - The pedigrees indicated here were obtained with...Ch. 11 - Approximately 3 of the population carries a mutant...Ch. 11 - The drug ivacaftor has recently been developed to...Ch. 11 - In the high-throughput DNA sequencing protocol...Ch. 11 - A researcher sequences the whole exome of a...Ch. 11 - As explained in the text, the cause of many...Ch. 11 - Figure 11.26 portrayed the analysis of Miller...Ch. 11 - A research paper published in the summer of 2012...Ch. 11 - Table 11.2 and Fig. 11.27 together portray the...Ch. 11 - The human RefSeq of the entire first exon of a...Ch. 11 - Mutations in the HPRT1 gene in humans result in at...Ch. 11 - Prob. 44P
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- How does the cost of food affect the nutritional choices people make?arrow_forwardBiopharmaceutics and Pharmacokinetics:Two-Compartment Model Zero-Order Absorption Questions SHOW ALL WORK, including equation used, variables used and each step to your solution, report your regression lines and axes names (with units if appropriate) :Calculate a-q a) B1, b) B2, c) hybrid rate constant (1) d) hybrid rate constant (2) e) t1/2,dist f) t1/2,elim g) k10 h) k12 i) k21 j) initial concentration (C0) k) central compartment volume (V1) l) steady-state volume (Vss) m) clearance (CL) AUC (0→10 min) using trapezoidal rule n) AUC (20→30 min) using trapezoidal rule o) AUCtail (AUC360→∞) p) total AUC (using short cut method) q) volume from AUC (VAUC)arrow_forwardWhat are some external influences that keep people from making healthy eating decisions?arrow_forward
- What type of structure(s) would you expect to see in peripheral membrane proteins? (mark all that apply) A. Amphipathic alpha helix (one side is hydrophilic and one side is hydrophobic) B. A hydrophobic beta barrel C. A hydrophobic alpha helix D. A chemical group attached to the protein that can anchor it to the membranearrow_forwardTemporal flexibility (the ability to change over time) of actin structures within a cell is maintained by… A. The growth/shrinkage cycle B. Periodic catastrophe C. GTP hydrolysis D. Treadmilling E. None of the abovearrow_forwardDuring in vitro polymerization of actin and microtubule filaments from their subunits, what causes the initial delay in filament growth? A.Nucleation B.Reaching homeostasis C.Nucleotide exchange D.ATP or GTP hydrolysis E.Treadmillingarrow_forward
- You expect to find which of the following in the Microtubule Organizing Center (MTOC)...(mark all that apply) A. Gamma tubulin B. XMAP215 C. Centrioles D. Kinesin-13arrow_forwardThe actin-nucleating protein formin has flexible “arms” containing binding sites that help recruit subunits in order to enhance microfilament polymerization. What protein binds these sites? A.Thymosin B.Profilin C.Cofilin D.Actin E.Tropomodulinarrow_forwardWhile investigating an unidentified motor protein, you discover that it has two heads that bind to actin. Based on this information, you could confidently determine that it is NOT... (mark all that apply) A. A myosin I motor B. A dynein motor C. A myosin VI motor D. A kinesin motorarrow_forward
- You isolate the plasma membrane of cells and find that . . . A. it contains regions with different lipid compositions B. it has different lipid types on the outer and cytosolic leaflets of the membrane C. neither are possible D. A and B both occurarrow_forwardYou are studying the mobility of a transmembrane protein that contains extracellular domains, one transmembrane domain, and a large cytosolic domain. Under normal conditions, this protein is confined to a particular region of the membrane due to the cortical actin cytoskeletal network. Which of the following changes is most likely to increase mobility of this protein beyond the normal restricted region of the membrane? A. Increased temperature B. Protease cleavage of the extracellular domain of the protein C. Binding to a free-floating extracellular ligand, such as a hormone D. Protease cleavage of the cytosolic domain of the protein E. Aggregation of the protein with other transmembrane proteinsarrow_forwardTopic: Benthic invertebrates as an indicator species for climate change, mapping changes in ecosystems (Historical Analysis & GIS) What objects or events has the team chosen to analyze? How does your team wish to delineate the domain or scale in which these objects or events operate? How does that limited domain facilitate a more feasible research project? What is your understanding of their relationships to other objects and events? Are you excluding other things from consideration which may influence the phenomena you seek to understand? Examples of such exclusions might include certain air-born pollutants; a general class of water bodies near Ottawa, or measurements recorded at other months of the year; interview participants from other organizations that are involved in the development of your central topic or issue. In what ways do your research questions follow as the most appropriate and/or most practical questions (given the circumstances) to pursue to better understand…arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
 Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Biology (MindTap Course List)
Biology
ISBN:9781337392938
Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. Berg
Publisher:Cengage Learning

Human Biology (MindTap Course List)
Biology
ISBN:9781305112100
Author:Cecie Starr, Beverly McMillan
Publisher:Cengage Learning

Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College


Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning
How to solve genetics probability problems; Author: Shomu's Biology;https://www.youtube.com/watch?v=R0yjfb1ooUs;License: Standard YouTube License, CC-BY
Beyond Mendelian Genetics: Complex Patterns of Inheritance; Author: Professor Dave Explains;https://www.youtube.com/watch?v=-EmvmBuK-B8;License: Standard YouTube License, CC-BY