
Concept explainers
Pigeons may exhibit a checkered or plain color pattern. In a series of controlled matings, the following data were obtained.
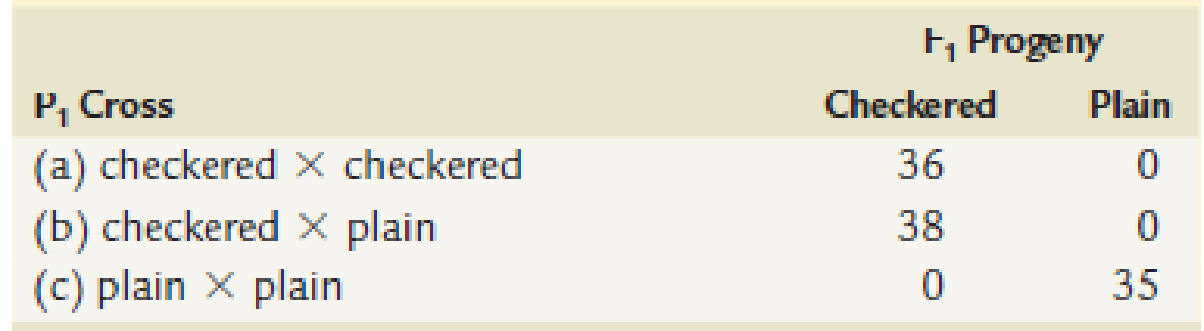
Then ⊢1 offspring were selectively mated with the following results. (The P1 cross giving rise to each ⊢1 pigeon is indicated in parentheses.)
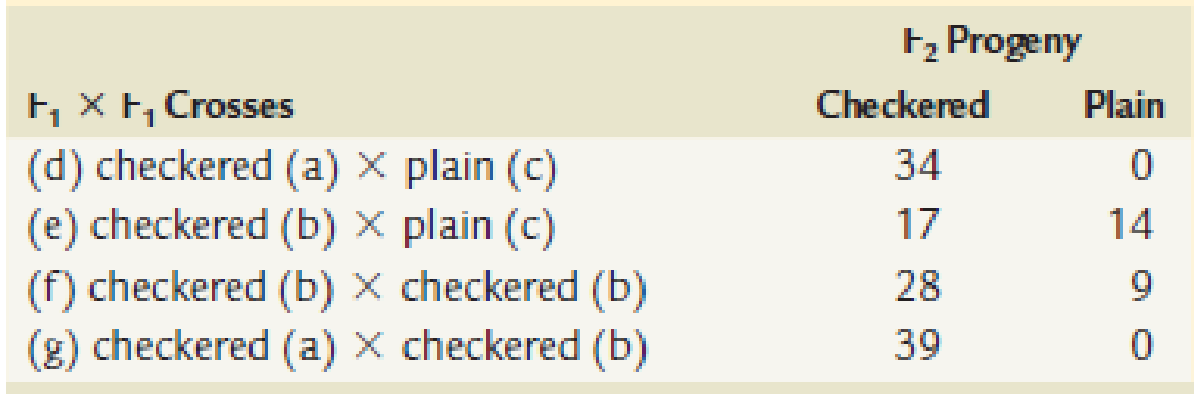
How are the checkered and plain patterns inherited? Select and assign symbols for the genes involved, and determine the genotypes of the parents and offspring in each cross.
To determine: The pattern of inheritance of checkered and plain pattern.
Introduction: Pattern of inheritance is defined as the pattern by which a particular trait or gene is transferred to the offspring and what are the conditions that is needed for its expression. There are five different types of pattern of inheritance namely X-linked dominant, X-linked recessive, autosomal recessive, autosomal dominant and mitochondrial inheritance.
Explanation of Solution
In the cross between checked and plain pattern shows a autosomal dominance inheritance in which checkered trait is a dominant trait and plain pattern is a recessive trait. This can be concluded because in a cross between checkered and plain pattern parents give all heterozygous offspring and since checkered trait being a dominant trait is expressed in all heterozygous progeny.
The inheritance pattern of offspring produced by crossing checkered and plain pattern parents is autosomal dominance inheritance.
To determine: The pattern of inheritance of checkered and plain pattern.
Introduction: Mendel gave three important postulates by observing the monohybrid crosses and Law of dominance is one of the three laws given by Mendel. According to this law a dominant trait expresses itself in both homozygous condition as well as heterozygous condition. On the contrary, the recessive trait is able to express only in homozygous condition.
Answer to Problem 1NST
Pictorial representation: Fig.1 cross showing the F1 and F2 progeny formed after crossing between checkered and plain pigeons.
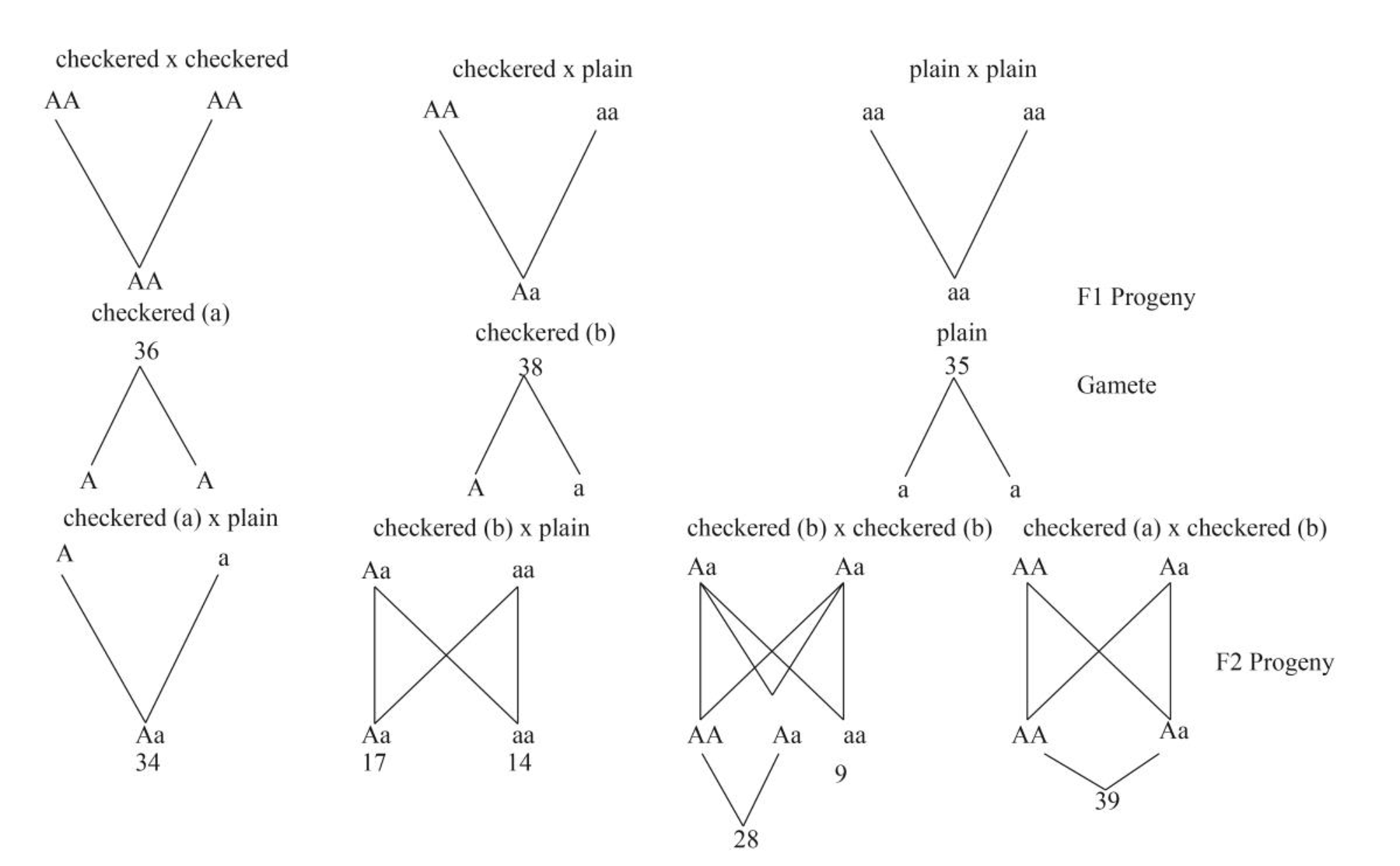
Fig.1: Crosses show the F1 and F2 progeny formed after crossing between checkered and plain pigeons.
Explanation of Solution
The dominant trait responsible for checkered pattern is represented by “A” and the recessive trait responsible for plain pattern is represented by “a”.
In the F2 cross when the F1 progeny is crossed with each other the cross between checkered (a) and plain gave 34 checkered and no plain progeny. When the checkered b progeny is crossed with the plain progeny 17 checkered and 14 plain progeny were formed. When checkered (b) is crossed with itself, 28 checkered and 9 non checkered progeny were formed and when checkered (a) and checkered (b) are crossed 39 checkered offspring’s were formed.
Want to see more full solutions like this?
Chapter 3 Solutions
Concepts of Genetics (12th Edition)
Additional Science Textbook Solutions
Genetic Analysis: An Integrated Approach (3rd Edition)
Human Anatomy & Physiology (2nd Edition)
Campbell Biology (11th Edition)
Organic Chemistry (8th Edition)
Human Physiology: An Integrated Approach (8th Edition)
Chemistry: An Introduction to General, Organic, and Biological Chemistry (13th Edition)
- What is this?arrow_forwardMolecular Biology A-C components of the question are corresponding to attached image labeled 1. D component of the question is corresponding to attached image labeled 2. For a eukaryotic mRNA, the sequences is as follows where AUGrepresents the start codon, the yellow is the Kozak sequence and (XXX) just represents any codonfor an amino acid (no stop codons here). G-cap and polyA tail are not shown A. How long is the peptide produced?B. What is the function (a sentence) of the UAA highlighted in blue?C. If the sequence highlighted in blue were changed from UAA to UAG, how would that affecttranslation? D. (1) The sequence highlighted in yellow above is moved to a new position indicated below. Howwould that affect translation? (2) How long would be the protein produced from this new mRNA? Thank youarrow_forwardMolecular Biology Question Explain why the cell doesn’t need 61 tRNAs (one for each codon). Please help. Thank youarrow_forward
- Molecular Biology You discover a disease causing mutation (indicated by the arrow) that alters splicing of its mRNA. This mutation (a base substitution in the splicing sequence) eliminates a 3’ splice site resulting in the inclusion of the second intron (I2) in the final mRNA. We are going to pretend that this intron is short having only 15 nucleotides (most introns are much longer so this is just to make things simple) with the following sequence shown below in bold. The ( ) indicate the reading frames in the exons; the included intron 2 sequences are in bold. A. Would you expected this change to be harmful? ExplainB. If you were to do gene therapy to fix this problem, briefly explain what type of gene therapy youwould use to correct this. Please help. Thank youarrow_forwardMolecular Biology Question Please help. Thank you Explain what is meant by the term “defective virus.” Explain how a defective virus is able to replicate.arrow_forwardMolecular Biology Explain why changing the codon GGG to GGA should not be harmful. Please help . Thank youarrow_forward
- Stage Percent Time in Hours Interphase .60 14.4 Prophase .20 4.8 Metaphase .10 2.4 Anaphase .06 1.44 Telophase .03 .72 Cytukinesis .01 .24 Can you summarize the results in the chart and explain which phases are faster and why the slower ones are slow?arrow_forwardCan you circle a cell in the different stages of mitosis? 1.prophase 2.metaphase 3.anaphase 4.telophase 5.cytokinesisarrow_forwardWhich microbe does not live part of its lifecycle outside humans? A. Toxoplasma gondii B. Cytomegalovirus C. Francisella tularensis D. Plasmodium falciparum explain your answer thoroughly.arrow_forward
- Select all of the following that the ablation (knockout) or ectopoic expression (gain of function) of Hox can contribute to. Another set of wings in the fruit fly, duplication of fingernails, ectopic ears in mice, excess feathers in duck/quail chimeras, and homeosis of segment 2 to jaw in Hox2a mutantsarrow_forwardSelect all of the following that changes in the MC1R gene can lead to: Changes in spots/stripes in lizards, changes in coat coloration in mice, ectopic ear formation in Siberian hamsters, and red hair in humansarrow_forwardPleiotropic genes are genes that (blank) Cause a swapping of organs/structures, are the result of duplicated sets of chromosomes, never produce protein products, and have more than one purpose/functionarrow_forward
 Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning





