
CAMPBELL BIOLOGY-W/MOD.MASTERBIOLOGY
11th Edition
ISBN: 9780134683461
Author: Urry
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 20, Problem 11TYU
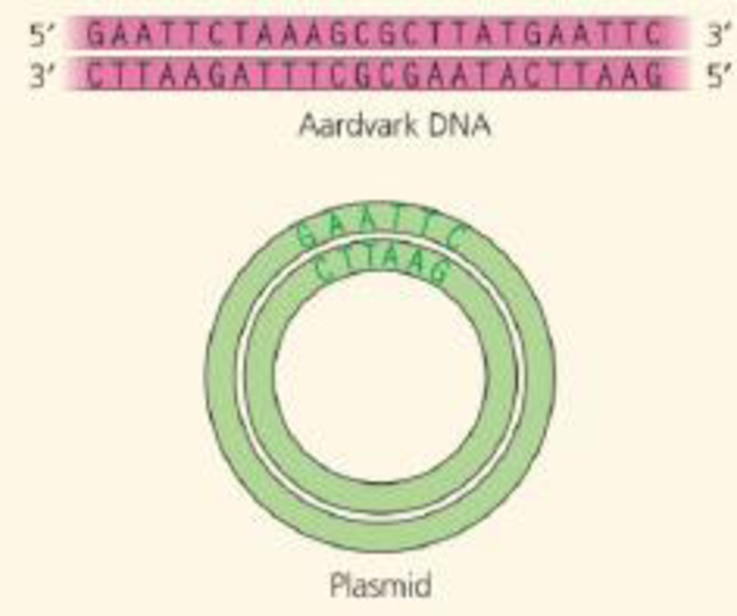
DRAW IT You are cloning an aardvark gene, using a bacterial plasmid as a vector. The green diagram shows the plasmid, which contains the restriction site for the enzyme used in Figure 20.5. Above the plasmid is a segment of linear aardvark DNA that was synthesized using PCR. Diagram your cloning procedure, and show what would happen to these two molecules during each step. Use one color for the aardvark DNA and its bases and another color for those of the plasmid. Label each step and all 5' and 3' ends.
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Diagram of check cell under low power and high power
a couple in which the father has the a blood type and the mother has the o blood type produce an offspring with the o blood type, how does this happen? how could two functionally O parents produce an offspring that has the a blood type?
What is the opening indicated by the pointer? (leaf x.s.)
stomate
guard cell
lenticel
intercellular space
none of these
Chapter 20 Solutions
CAMPBELL BIOLOGY-W/MOD.MASTERBIOLOGY
Ch. 20.1 - Prob. 1CCCh. 20.1 - DRAW IT One Strand of a DNA molecule has the...Ch. 20.1 - What are some potential difficulties in using...Ch. 20.1 - VISUAL SKILLS Compare Figure 20.7 with Figure...Ch. 20.2 - Prob. 1CCCh. 20.2 - Prob. 2CCCh. 20.3 - Based on current knowledge, how would you explain...Ch. 20.3 - Prob. 2CCCh. 20.3 - Prob. 3CCCh. 20.4 - What is the advantage of using stem cells for gene...
Ch. 20.4 - Prob. 2CCCh. 20.4 - Prob. 3CCCh. 20 - Describe how the process of gene doning results in...Ch. 20 - What useful Information is obtained by detecting...Ch. 20 - Describe how, using mice. a researcher could carry...Ch. 20 - What factors affecf whether a given genetic...Ch. 20 - In DNA technology, the term vector can refer to...Ch. 20 - Which of the following tools of DNA technology is...Ch. 20 - Prob. 3TYUCh. 20 - A paleontologist has recovered a bit of tissue...Ch. 20 - DNA technology has many medical applications....Ch. 20 - Which of the following is not true of cDNA...Ch. 20 - Expression of a cloned eukaryotic gene in a...Ch. 20 - Which Ii of the following sequences in...Ch. 20 - Prob. 9TYUCh. 20 - MAKE CONNECTIONS Looking at Figure 20.15, what...Ch. 20 - DRAW IT You are cloning an aardvark gene, using a...Ch. 20 - EVOLUTlON CONNECTION Ethical considerations aside,...Ch. 20 - Prob. 13TYUCh. 20 - Prob. 14TYUCh. 20 - The water in the Yellowstone National Park hot...
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Identify the indicated tissue? (stem x.s.) parenchyma collenchyma sclerenchyma ○ xylem ○ phloem none of thesearrow_forwardWhere did this structure originate from? (Salix branch root) epidermis cortex endodermis pericycle vascular cylinderarrow_forwardIdentify the indicated tissue. (Tilia stem x.s.) parenchyma collenchyma sclerenchyma xylem phloem none of thesearrow_forward
- Identify the indicated structure. (Cucurbita stem l.s.) pit lenticel stomate tendril none of thesearrow_forwardIdentify the specific cell? (Zebrina leaf peel) vessel element sieve element companion cell tracheid guard cell subsidiary cell none of thesearrow_forwardWhat type of cells flank the opening on either side? (leaf x.s.) vessel elements sieve elements companion cells tracheids guard cells none of thesearrow_forward
- What specific cell is indicated. (Cucurbita stem I.s.) vessel element sieve element O companion cell tracheid guard cell none of thesearrow_forwardWhat specific cell is indicated? (Aristolochia stem x.s.) vessel element sieve element ○ companion cell O O O O O tracheid O guard cell none of thesearrow_forwardIdentify the tissue. parenchyma collenchyma sclerenchyma ○ xylem O phloem O none of thesearrow_forward
- Please answer q3arrow_forwardRespond to the following in a minimum of 175 words: How might CRISPR-Cas 9 be used in research or, eventually, therapeutically in patients? What are some potential ethical issues associated with using this technology? Do the advantages of using this technology outweigh the disadvantages (or vice versa)? Explain your position.arrow_forwardYou are studying the effect of directional selection on body height in three populations (graphs a, b, and c below). (a) What is the selection differential? Show your calculation. (2 pts) (b) Which population has the highest narrow sense heritability for height? Explain your answer. (2 pts) (c) If you examined the offspring in the next generation in each population, which population would have the highest mean height? Why? (2 pts) (a) Midoffspring height (average height of offspring) Short Short Short Short (c) Short (b) Short Tall Short Tall Short Short Tall Midparent height (average height of Mean of population = 65 inches Mean of breading parents = 70 inches Mean of population = 65 inches Mean of breading parents = 70 inches Mean of population = 65 inches Mean of breading parents = 70 inchesarrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
 Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning


Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College
Molecular Techniques: Basic Concepts; Author: Dr. A's Clinical Lab Videos;https://www.youtube.com/watch?v=7HFHZy8h6z0;License: Standard Youtube License