
Genetics: From Genes to Genomes
6th Edition
ISBN: 9781259700903
Author: Leland Hartwell Dr., Michael L. Goldberg Professor Dr., Janice Fischer, Leroy Hood Dr.
Publisher: McGraw-Hill Education
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 11, Problem 22P
Microarrays were used to determine the genotypes of seven embryos (made by in vitro fertilization) with regard to sickle-cell anemia. Each pair of squares in the accompanying figure represents two ASOs, one specific for the HbβA allele (A) and the other for the HBβ allele (T), attached to a chip of silicon and hybridized with fluorescently labeled PCR product from a single cell from one of the embryos. The hybridizations were performed at three different temperatures (80°C, 60°C, and 40°C) as shown.
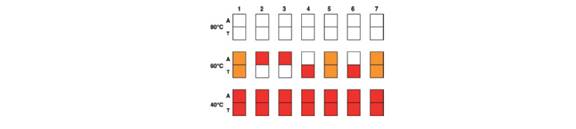
| a. | Why do you think the PCR step is needed for this microarray analysis? |
| b. | Make a sketch of the location in genomic DNA of the PCR primers relative to the sickle-cell mutation. Indicate the 5′-to-3′ polarities of all DNA molecules involved. |
| c. | Why is no hybridization seen at 80°C? |
| d. | Why do you see strong hybridization of all genomic DNA probes to both ASOs at 40°C? |
| e. | What are the genotypes of the seven embryos? Which of these embryos would you choose to implant into the mother’s uterus to avoid the possibility that the child would have sickle-cell anemia? |
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Explain in a small summary how:
What genetic information can be obtained from a Punnet square? What genetic information cannot be determined from a Punnet square?
Why might a Punnet Square be beneficial to understanding genetics/inheritance?
In a small summary write down:
Not part of a graded assignment, from a past midterm
Chapter 11 Solutions
Genetics: From Genes to Genomes
Ch. 11 - Choose the phrase from the right column that best...Ch. 11 - Would you characterize the pattern of inheritance...Ch. 11 - Would you be more likely to find single nucleotide...Ch. 11 - A recent estimate of the rate of base...Ch. 11 - If you examine Fig. 11.5 closely, you will note...Ch. 11 - Approximately 50 million SNPs have thus far been...Ch. 11 - Mutations at simple sequence repeat SSR loci occur...Ch. 11 - Humans and gorillas last shared a common ancestor...Ch. 11 - In 2015, an international team of scientists...Ch. 11 - Using PCR, you want to amplify an approximately 1...
Ch. 11 - Prob. 11PCh. 11 - The previous problem raises several interesting...Ch. 11 - You want to make a recombinant DNA in which a PCR...Ch. 11 - You sequence a PCR product amplified from a...Ch. 11 - Prob. 15PCh. 11 - The trinucleotide repeat region of the Huntington...Ch. 11 - Sperm samples were taken from two men just...Ch. 11 - Prob. 18PCh. 11 - a. It is possible to perform DNA fingerprinting...Ch. 11 - On July 17, 1918, Tsar Nicholas II; his wife the...Ch. 11 - The figure that follows shows DNA fingerprint...Ch. 11 - Microarrays were used to determine the genotypes...Ch. 11 - A partial sequence of the wild-type HbA allele is...Ch. 11 - a. In Fig. 11.17b, PCR is performed to amplify...Ch. 11 - The following figure shows a partial microarray...Ch. 11 - Scientists were surprised to discover recently...Ch. 11 - The microarray shown in Problem 25 analyzes...Ch. 11 - The figure that follows shows the pedigree of a...Ch. 11 - One of the difficulties faced by human geneticists...Ch. 11 - Now consider a mating between consanguineous...Ch. 11 - The pedigree shown in Fig. 11.22 was crucial to...Ch. 11 - You have identified a SNP marker that in one large...Ch. 11 - The pedigrees indicated here were obtained with...Ch. 11 - Approximately 3 of the population carries a mutant...Ch. 11 - The drug ivacaftor has recently been developed to...Ch. 11 - In the high-throughput DNA sequencing protocol...Ch. 11 - A researcher sequences the whole exome of a...Ch. 11 - As explained in the text, the cause of many...Ch. 11 - Figure 11.26 portrayed the analysis of Miller...Ch. 11 - A research paper published in the summer of 2012...Ch. 11 - Table 11.2 and Fig. 11.27 together portray the...Ch. 11 - The human RefSeq of the entire first exon of a...Ch. 11 - Mutations in the HPRT1 gene in humans result in at...Ch. 11 - Prob. 44P
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Noggin mutation: The mouse, one of the phenotypic consequences of Noggin mutationis mispatterning of the spinal cord, in the posterior region of the mouse embryo, suchthat in the hindlimb region the more ventral fates are lost, and the dorsal Pax3 domain isexpanded. (this experiment is not in the lectures).a. Hypothesis for why: What would be your hypothesis for why the ventral fatesare lost and dorsal fates expanded? Include in your answer the words notochord,BMP, SHH and either (or both of) surface ectoderm or lateral plate mesodermarrow_forwardNot part of a graded assignment, from a past midtermarrow_forwardNot part of a graded assignment, from a past midtermarrow_forward
- please helparrow_forwardWhat does the heavy dark line along collecting duct tell us about water reabsorption in this individual at this time? What does the heavy dark line along collecting duct tell us about ADH secretion in this individual at this time?arrow_forwardBiology grade 10 study guidearrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage Learning
Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College


Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Case Studies In Health Information Management
Biology
ISBN:9781337676908
Author:SCHNERING
Publisher:Cengage

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning

Principles Of Radiographic Imaging: An Art And A ...
Health & Nutrition
ISBN:9781337711067
Author:Richard R. Carlton, Arlene M. Adler, Vesna Balac
Publisher:Cengage Learning

Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College
Molecular Techniques: Basic Concepts; Author: Dr. A's Clinical Lab Videos;https://www.youtube.com/watch?v=7HFHZy8h6z0;License: Standard Youtube License