
Concept explainers
RIPs as Cancer Drugs Researchers are taking a page from the structure-function relationship of RIPs in their quest for cancer treatments. The most toxic RIPs, remember, have one domain that interferes with ribosomes, and another that carries them into cells. Melissa Cheung and her colleagues incorporated a peptide that binds to skin cancer cells into the enzymatic part of an RIP, the E. coli Shiga-like toxin. The researchers created a new RIP that specifically kills .skin cancer cells, which are notoriously resistant to established therapies. Some of their results are shown in FIGURE 9.17.
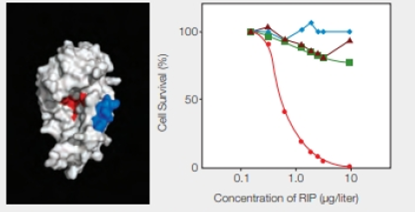
FIGURE 9.17 Effect of an engineered RIP on cancer cells. The model on the left shows the enzyme portion of E. coli Shiga-like toxin engineered to carry a small sequence of amino acids (in blue) that targets skin cancer cells. (Red indicates the active site.) The graph on the right shows the effect of this engineered RIP on human cancer cells of the skin ( ); breast (
); breast ( ) liver (
) liver ( ); and prostate (
); and prostate ( ).
).
Which cells had the greatest response to an increase in concentration of the engineered RIP?
To determine: The type of cells that had the greatest response to an increase in the concentration of the engineered RIP.
Introduction: Ribosome-inactivating proteins (RIPs) inactivate the ribosomes and prevent protein synthesis in a cell. The toxic RIPs have a domain that makes them enter into the cell and another domain that interferes with the ribosome. They have antiviral and anticancer properties and are used to design drugs for HIV and cancer.
Answer to Problem 1DAA
Correct answer: The greatest response in the form of fall in cell’s survival percentage with an increase in the concentration of engineered RIP is seen in the skin cancer cells.
Explanation of Solution
As given in the problem statement, Researcher M and her colleagues incorporated a peptide into the enzymatic part of a RIP, the E. coli Shiga-like toxin. The peptide specifically binds to the skin cancer cells, and thus, the newly synthesized RIP kills the skin cancer cells.
Refer Fig. 9.17, “Effect of an engineered RIP on cancer cells”, in the textbook. The model shown on the left indicates a blue-colored enzyme region of E. coli Shiga-like toxin that is engineered to carry the peptide sequence specific for the skin cancer cells. The red color indicates the active site of RIP.
The graphical representation that is shown in Fig. 9.17 on the right side indicates the effect of the engineered RIP on different human cancer cells indicated by different colors and shapes. They include skin, breast, liver, and prostate cancer cells with red, blue, brown, and green color, respectively. The concentration of RIP (µg/liter) is plotted with the percentage of cell survival. As shown in the graph, as the concentration of RIP increases, there is a significant drop in the skin cancer cells percentage. It reaches to zero at RIP concentration of 10 µg/liter. In the case of the other cancer cells, there is lesser variability.
Thus, the greatest response in the form of fall in cell’s survival percentage with an increase in the concentration of engineered RIP is seen in the skin cancer cells.
Want to see more full solutions like this?
Chapter 9 Solutions
Biology: The Unity and Diversity of Life (MindTap Course List)
Additional Science Textbook Solutions
Fundamentals of Physics Extended
Laboratory Manual For Human Anatomy & Physiology
SEELEY'S ANATOMY+PHYSIOLOGY
Microbiology Fundamentals: A Clinical Approach
Human Physiology: An Integrated Approach (8th Edition)
- You intend to insert patched dominant negative DNA into the left half of the neural tube of a chick. 1) Which side of the neural tube would you put the positive electrode to ensure that the DNA ends up on the left side? 2) What would be the internal (within the embryo) control for this experiment? 3) How can you be sure that the electroporation method itself is not impacting the embryo? 4) What would you do to ensure that the electroporation is working? How can you tell?arrow_forwardDescribe a method to document the diffusion path and gradient of Sonic Hedgehog through the chicken embryo. If modifying the protein, what is one thing you have to consider in regards to maintaining the protein’s function?arrow_forwardThe following table is from Kumar et. al. Highly Selective Dopamine D3 Receptor (DR) Antagonists and Partial Agonists Based on Eticlopride and the D3R Crystal Structure: New Leads for Opioid Dependence Treatment. J. Med Chem 2016.arrow_forward
- The following figure is from Caterina et al. The capsaicin receptor: a heat activated ion channel in the pain pathway. Nature, 1997. Black boxes indicate capsaicin, white circles indicate resinferatoxin. You are a chef in a fancy new science-themed restaurant. You have a recipe that calls for 1 teaspoon of resinferatoxin, but you feel uncomfortable serving foods with "toxins" in them. How much capsaicin could you substitute instead?arrow_forwardWhat protein is necessary for packaging acetylcholine into synaptic vesicles?arrow_forward1. Match each vocabulary term to its best descriptor A. affinity B. efficacy C. inert D. mimic E. how drugs move through body F. how drugs bind Kd Bmax Agonist Antagonist Pharmacokinetics Pharmacodynamicsarrow_forward
- 50 mg dose of a drug is given orally to a patient. The bioavailability of the drug is 0.2. What is the volume of distribution of the drug if the plasma concentration is 1 mg/L? Be sure to provide units.arrow_forwardDetermine Kd and Bmax from the following Scatchard plot. Make sure to include units.arrow_forwardChoose a catecholamine neurotransmitter and describe/draw the components of the synapse important for its signaling including synthesis, packaging into vesicles, receptors, transporters/degradative enzymes. Describe 2 drugs that can act on this system.arrow_forward
- The following figure is from Caterina et al. The capsaicin receptor: a heat activated ion channel in the pain pathway. Nature, 1997. Black boxes indicate capsaicin, white circles indicate resinferatoxin. a) Which has a higher potency? b) Which is has a higher efficacy? c) What is the approximate Kd of capsaicin in uM? (you can round to the nearest power of 10)arrow_forwardWhat is the rate-limiting-step for serotonin synthesis?arrow_forwardWhat enzyme is necessary for synthesis of all of the monoamines?arrow_forward
 Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning





