
Concept explainers
RIPs as Cancer Drugs Researchers are taking a page from the structure-function relationship of RIPs in their quest for cancer treatments. The most toxic RIPs, remember, have one domain that interferes with ribosomes, and another that carries them into cells. Melissa Cheung and her colleagues incorporated a peptide that binds to skin cancer cells into the enzymatic part of an RIP, the E. coli Shiga-like toxin. The researchers created a new RIP that specifically kills skin cancer cells, which are notoriously resistant to established therapies. Some of their results are shown in FIGURE 9.17.
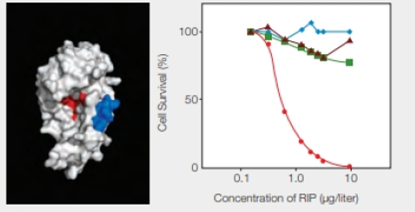
FIGURE 9.17 Effect of an engineered RIP on cancer cells. The model on the left shows the enzyme portion of E. coli Shiga-like toxin engineered to carry a small sequence of amino acids (in blue) that targets skin cancer cells. (Red indicates the active site.) The graph on the right shows the effect of this engineered RIP on human cancer cells of the skin ( ); breast (
); breast ( ) liver (
) liver ( ); and prostate (
); and prostate ( ).
).
Why are some of the data points linked by curved lines?
Want to see the full answer?
Check out a sample textbook solution
Chapter 9 Solutions
Biology: The Unity and Diversity of Life (MindTap Course List)
- Identify the indicated cavity (Fucus). a. antheridia b. conceptacel c. receptacle d. oogonium e. none of thesearrow_forwardIdentify the indicated structure (Saprolegnia). a. antheridium O b. oospore c.sperm d. auxospore e. tetraspore Of. zygosporearrow_forwardUsing information from the primary literature (several references have been provided as a starting point below) please answer the following question: Based on your review of the literature on rewilding, what are the major scientific pros and cons for rewilding? Please note that the focus of this assignment are the (biological) scientific issues associated with rewilding. As will be discussed in class, there are a number of non-scientific issues involved or implicated in rewilding, all ultimately affecting the public acceptability of rewilding. Although these issues are important – indeed, critical – in this assignment you should focus on the biological science issues and questions. Details: You must enumerate at least two pros and at least two cons. Your answer should be no more than 500 well-chosen words, excluding references. Think carefully about how best to organize and structure your answer. Aim for high information density: say a lot, but say it succinctly. Recall Nietzche’s…arrow_forward
- Using information from the primary literature (several references have been provided as a starting point below) please answer the following question: Based on your review of the literature on rewilding, what are the major scientific pros and cons for rewilding? Please note that the focus of this assignment are the (biological) scientific issues associated with rewilding. As will be discussed in class, there are a number of non-scientific issues involved or implicated in rewilding, all ultimately affecting the public acceptability of rewilding. Although these issues are important – indeed, critical – in this assignment you should focus on the biological science issues and questions. Details: You must enumerate at least two pros and at least two cons. Your answer should be no more than 500 well-chosen words, excluding references. Think carefully about how best to organize and structure your answer. Aim for high information density: say a lot, but say it succinctly. Recall Nietzche’s…arrow_forwardNow draw a rough sketch of what the control data might look like if in addition to the specific binding, there was also a considerable amount of nonspecific binding (again using a normal dose/response curve) (do % total bound ligand vs concentration)arrow_forwardWhat are functions of cuboidal cells in the kidney? Select all that apply. Concentration of gases Dilution of chemicals Secretion of molecules Nutrition to tissues Support of tissues Absorption of moleculesarrow_forward
- question1 In plants, epithelial tissue is only found as the outermost cell layer and acts as a barrier. In humans, epithelial tissue is found inside the body as well as on the surface. What function(s) does/do epithelial tissue carry out in humans? Select all that apply. Waste storage Filtration Oxygen transport Protection Diffusion Osmosis Absorptionarrow_forwardWhat words best describes this organism? a. Unicellular/nonmotile Ob. unicellular/motile c. colonial/nonmotile d. colonial/motile e. multicelluar O f. siphonous g. none of thesearrow_forwardIdentify the phylum or class. a. Euglenophyta b. Dinoflagellata c. Bacillariophyceae d. Oomycetes e. Phaeophyceae O f. Myxomycota g. Xanthophyceae ○ h. Chrysophyceae i. Dictyosteliomycota O j. Rhodophyta Ok. Chlorophyceaens I. Charophyceaensarrow_forward
 Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning





