
Concept explainers
RIPs as Cancer Drugs Researchers are taking a page from the structure-function relationship of RIPs in their quest for cancer treatments. The most toxic RIPs, remember, have one domain that interferes with ribosomes, and another that carries them into cells. Melissa Cheung and her colleagues incorporated a peptide that binds to skin cancer cells into the enzymatic part of an RIP, the E. coli Shiga-like toxin. The researchers created a new RIP that specifically kills .skin cancer cells, which are notoriously resistant to established therapies. Some of their results are shown in FIGURE 9.17.
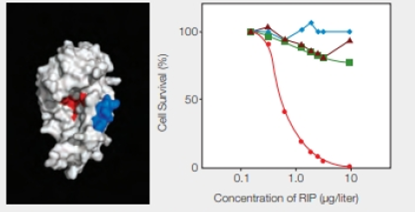
FIGURE 9.17 Effect of an engineered RIP on cancer cells. The model on the left shows the enzyme portion of E. coli Shiga-like toxin engineered to carry a small sequence of amino acids (in blue) that targets skin cancer cells. (Red indicates the active site.) The graph on the right shows the effect of this engineered RIP on human cancer cells of the skin ( ); breast (
); breast ( ) liver (
) liver ( ); and prostate (
); and prostate ( ).
).
Which cells had the greatest response to an increase in concentration of the engineered RIP?
To determine: The type of cells that had the greatest response to an increase in the concentration of the engineered RIP.
Introduction: Ribosome-inactivating proteins (RIPs) inactivate the ribosomes and prevent protein synthesis in a cell. The toxic RIPs have a domain that makes them enter into the cell and another domain that interferes with the ribosome. They have antiviral and anticancer properties and are used to design drugs for HIV and cancer.
Answer to Problem 1DAA
Correct answer: The greatest response in the form of fall in cell’s survival percentage with an increase in the concentration of engineered RIP is seen in the skin cancer cells.
Explanation of Solution
As given in the problem statement, Researcher M and her colleagues incorporated a peptide into the enzymatic part of a RIP, the E. coli Shiga-like toxin. The peptide specifically binds to the skin cancer cells, and thus, the newly synthesized RIP kills the skin cancer cells.
Refer Fig. 9.17, “Effect of an engineered RIP on cancer cells”, in the textbook. The model shown on the left indicates a blue-colored enzyme region of E. coli Shiga-like toxin that is engineered to carry the peptide sequence specific for the skin cancer cells. The red color indicates the active site of RIP.
The graphical representation that is shown in Fig. 9.17 on the right side indicates the effect of the engineered RIP on different human cancer cells indicated by different colors and shapes. They include skin, breast, liver, and prostate cancer cells with red, blue, brown, and green color, respectively. The concentration of RIP (µg/liter) is plotted with the percentage of cell survival. As shown in the graph, as the concentration of RIP increases, there is a significant drop in the skin cancer cells percentage. It reaches to zero at RIP concentration of 10 µg/liter. In the case of the other cancer cells, there is lesser variability.
Thus, the greatest response in the form of fall in cell’s survival percentage with an increase in the concentration of engineered RIP is seen in the skin cancer cells.
Want to see more full solutions like this?
Chapter 9 Solutions
Biology: The Unity and Diversity of Life (MindTap Course List)
Additional Science Textbook Solutions
Fundamentals of Physics Extended
Laboratory Manual For Human Anatomy & Physiology
SEELEY'S ANATOMY+PHYSIOLOGY
Microbiology Fundamentals: A Clinical Approach
Human Physiology: An Integrated Approach (8th Edition)
- What is behavioral adaptarrow_forward22. Which of the following mutant proteins is expected to have a dominant negative effect when over- expressed in normal cells? a. mutant PI3-kinase that lacks the SH2 domain but retains the kinase function b. mutant Grb2 protein that cannot bind to RTK c. mutant RTK that lacks the extracellular domain d. mutant PDK that has the PH domain but lost the kinase function e. all of the abovearrow_forwardWhat is the label ?arrow_forward
- Can you described the image? Can you explain the question as well their answer and how to get to an answer to an problem like this?arrow_forwardglg 112 mid unit assignment Identifying melting processesarrow_forwardGive only the mode of inheritance consistent with all three pedigrees and only two reasons that support this, nothing more, (it shouldn't take too long)arrow_forward
- Oarrow_forwardDescribe the principle of homeostasis.arrow_forwardExplain how the hormones of the glands listed below travel around the body to target organs and tissues : Pituitary gland Hypothalamus Thyroid Parathyroid Adrenal Pineal Pancreas(islets of langerhans) Gonads (testes and ovaries) Placentaarrow_forward
 Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning





