
Concept explainers
Data Analysis Activities
Neanderthal Hair Color The MC1R gene regulates pigmentation in humans (Sections 13.5 and 14.2), so Loss-of-function mutations in this gene affect hair and skin color. A person with two mutated alleles for this gene makes more of the reddish melanin than the brownish melanin, resu.lti.ng in red hair and pale skin. DNA extracted from two Neanderthal fossils contains a mutated MC1R allele that has not yet been found in humans. To see how the Neanderthal mutation affects the function of the MC1R gene, Carles Lalueza-Fox and her team introduced the allele in to cultured monkey cells (FIGURE 26.12).
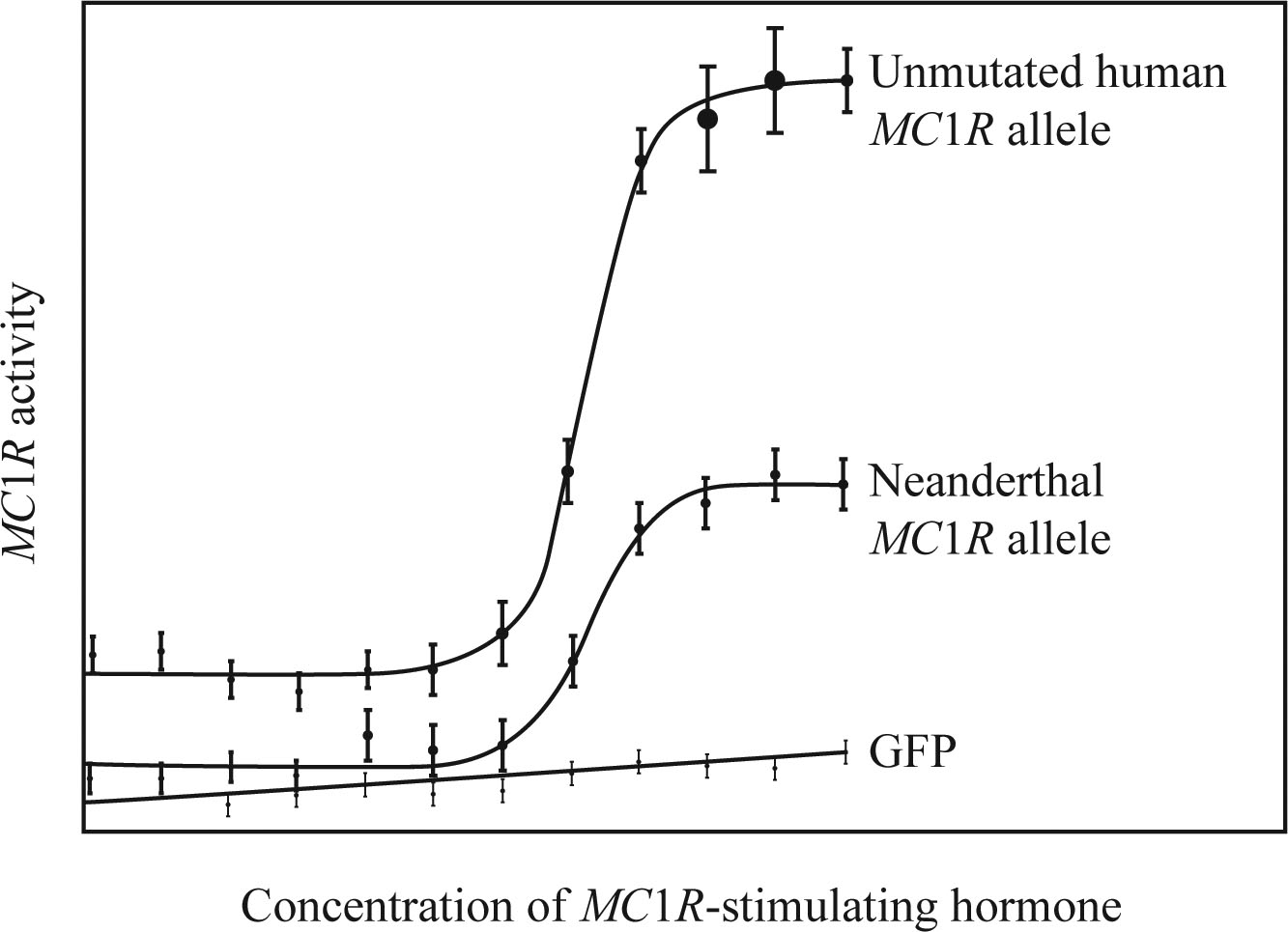
FIGURE 26.12 MC1Ractivity. Activity is shown in monkey cells transgenic for an unmutated MC1R gene, the Neanderthal MC1R allele, or the gene for green fluorescent protein (GFP). GFP is not related to MC1R.
How did MCR1 activity in monkey cells with the mutant allele differ from that in cells with the normal allele?
To explain: The way in which the MC1R activity in the monkey cells with the mutant allele differs from that in cells with the normal allele.
Introduction: Neanderthals are extinct hominins that are the closest relatives of Homo sapiens and habituated to the Middle East, Europe, and Asia. The pigmentation in humans is regulated by MC1R. The loss of function mutation in the MC1R gene, in turn, affects the skin and hair color. The homozygous mutation of the gene results in red hair and pale skin phenotypes.
Explanation of Solution
Scientist C and her team worked on MC1R mutated form that was present only in the Neanderthals. The team incorporated the allele from Neanderthal into cultured monkey cells.
Based on the results from the experiment, a graph was plotted with the concentration of the hormone that stimulates MC1R in the X-axis and the activity of MC1R in the Y-axis. Three scatter plots were plotted: one for green fluorescent protein (GFP), one for Neanderthal MC1R allele, and one for unmutated human MC1R allele. The activity of MC1R was very high in the transgenic monkey cells when compared to the normal allele.
The MC1R activity in the monkey cells with the mutant allele was much greater than the cells that had a normal allele for MC1R.
Want to see more full solutions like this?
Chapter 26 Solutions
Biology: The Unity and Diversity of Life (MindTap Course List)
Additional Science Textbook Solutions
Biological Science (6th Edition)
Laboratory Manual For Human Anatomy & Physiology
SEELEY'S ANATOMY+PHYSIOLOGY
Biology: Life on Earth with Physiology (11th Edition)
Chemistry: A Molecular Approach (4th Edition)
- What is a gain of function mutation?arrow_forwardMolecular Biology Question: Please help. Thank you Is Southern hybridization's purpose detecting specific nucleotide sequences? How so?arrow_forwardUse the following information to answer the question(s) below. Martin Wikelski and L. Michael Romero (Body size, performance and fitness in Galápagos marine iguanas, Integrative and Comparative Biology 43 [2003]:376-86) measured the snout-to-vent (anus) length of Galápagos marine iguanas and observed the percent survival of different-sized animals, all of the same age. The graph shows the log snout-vent length (SVL, a measure of overall body size) plotted against the percent survival of these different size classes for males and females. Survival (%) 100- 80- 60- 40- 20- 0+ 1.9 T 2 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Log SVL (mm) 19) Examine the figure above. What type of selection for body size appears to be occurring in these marine iguanas? A) directional selection B) stabilizing selection C) disruptive selection D) You cannot determine the type of selection from the above information. 3arrow_forward
- 24) Use the following information to answer the question below. Researchers studying a small milkweed population note that some plants produce a toxin and other plants do not. They identify the gene responsible for toxin production. The dominant allele (T) codes for an enzyme that makes the toxin, and the recessive allele (t) codes for a nonfunctional enzyme that cannot produce the toxin. Heterozygotes produce an intermediate amount of toxin. The genotypes of all individuals in the population are determined (see table) and used to determine the actual allele frequencies in the population. TT 0.49 Tt 0.42 tt 0.09 Refer to the table above. Is this population in Hardy-Weinberg equilibrium? A) Yes. C) No; there are more homozygotes than expected. B) No; there are more heterozygotes than expected. D) It is impossible to tell.arrow_forward30) A B CDEFG Refer to the accompanying figure. Which of the following forms a monophyletic group? A) A, B, C, and D B) C and D C) D, E, and F D) E, F, and Garrow_forwardMolecular Biology Question. Please help with step solution and explanation. Thank you: The Polymerase Chain Reaction (PCR) reaction consists of three steps denaturation, hybridization, and elongation. Please describe what occurs in the annealing step of the PCR reaction. (I think annealing step is hybridization). What are the other two steps of PCR, and what are their functions? Next, suppose the Tm for the two primers being used are 54C for Primer A and 67C for Primer B. Regarding annealing step temperature, I have the following choices for the temperature used during the annealing step:(a) 43C (b) 49C (c) 62C (d) 73C Which temperature/temperatures should I choose? What is the corresponding correct explanation, and why would I not use the other temperatures? Have a good day!arrow_forward
- Using the data provided on the mean body mass and horn size of 4-year-old male sheep, draw a scatterplot graph to examine how body mass and horn size changed over time.arrow_forwardPlease write a 500-word report about the intake of saturated fat, sodium, alcoholic beverages, or added sugar in America. Choose ONE of these and write about what is recommended by the Dietary Guidelines for Americans (guideline #4) and why Americans exceed the intake of that nutrient. Explain what we could do as a society and/or individuals to reduce our intake of your chosen nutrient.arrow_forwardWrite a 500-word report indicating how you can change the quantity or quality of TWO nutrients where your intake was LOWER than what is recommended by the Dietary Guidelines for Americans and/or the DRIs. Indicate how the lack of the nutrient may affect your health. For full credit, all of the following points must be addressed and elaborated on in more detail for each nutrient: The name of the nutrient At least 2 main functions of the nutrient (example: “Vitamin D regulates calcium levels in the blood and calcification of bones.”) Your percent intake compared to the RDA/DRI (example “I consumed 50% of the RDA for vitamin D”) Indicate why your intake was below the recommendations (example: “I only had one serving of dairy products and that was why I was below the recommendations for vitamin D”) How would you change your dietary pattern to meet the recommendations? – be sure to list specific foods (example: “I would add a yogurt and a glass of milk to each day in order to increase my…arrow_forward
- Why are nutrient absorption and dosage levels important when taking multivitamins and vitamin and mineral supplements?arrow_forwardI'm struggling with this topic and would really appreciate your help. I need to hand-draw a diagram and explain the process of sexual differentiation in humans, including structures, hormones, enzymes, and other details. Could you also make sure to include these terms in the explanation? . Gonads . Wolffian ducts • Müllerian ducts . ⚫ Testes . Testosterone • Anti-Müllerian Hormone (AMH) . Epididymis • Vas deferens ⚫ Seminal vesicles ⚫ 5-alpha reductase ⚫ DHT - Penis . Scrotum . Ovaries • Uterus ⚫ Fallopian tubes - Vagina - Clitoris . Labia Thank you so much for your help!arrow_forwardRequisition Exercise A phlebotomist goes to a patient’s room with the following requisition. Hometown Hospital USA 125 Goodcare Avenue Small Town, USAarrow_forward
 Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning





