
Biology
12th Edition
ISBN: 9781260494570
Author: Raven, Peter
Publisher: MCGRAW-HILL HIGHER EDUCATION
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 18, Problem 1S
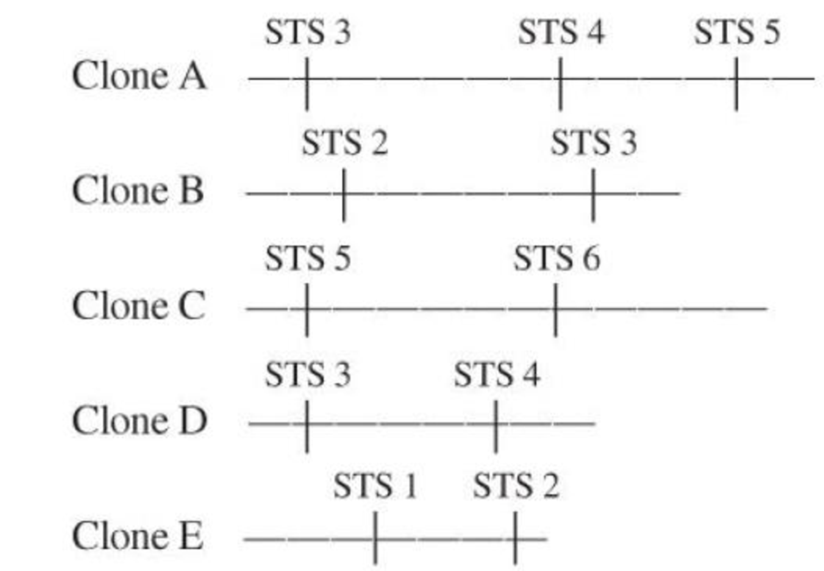
You are in the early stages of a genome-sequencing project. You have isolated a number of clones from a bacterial artificial chromosome (BAC) library and mapped the inserts in these clones using STSs. Use the STSs shown to align the clones into a contiguous sequence of the genome (a contig).
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Molecular Biology
Please help with question. thank you
You are studying the expression of the lac operon. You have isolated mutants as described below. In the presence of glucose, explain/describe what would happen, for each mutant, to the expression of the lac operon when you add lactose AND what would happen when the bacteria has used up all of the lactose (if the mutant is able to use lactose).5. Mutations in the lac operator that strengthen the binding of the lac repressor 200 fold
6. Mutations in the promoter that prevent binding of RNA polymerase
7. Mutations in CRP/CAP protein that prevent binding of cAMP8. Mutations in sigma factor that prevent binding of sigma to core RNA polymerase
Molecular Biology
Please help and there is an attached image. Thank you.
A bacteria has a gene whose protein/enzyme product is involved with the synthesis of a lipid necessary for the synthesis of the cell membrane. Expression of this gene requires the binding of a protein (called ACT) to a control sequence (called INC) next to the promoter.
A. Is the expression/regulation of this gene an example of induction or repression?Please explain:B. Is this expression/regulation an example of positive or negative control?C. When the lipid is supplied in the media, the expression of the enzyme is turned off.Describe one likely mechanism for how this “turn off” is accomplished.
Molecular Biology
Please help. Thank you.
Discuss/define the following:(a) poly A polymerase (b) trans-splicing (c) operon
Chapter 18 Solutions
Biology
Ch. 18.1 - Prob. 1LOCh. 18.1 - Describe the pros and cons of restriction mapping,...Ch. 18.1 - Prob. 3LOCh. 18.2 - Discriminate between dideoxy terminator sequencing...Ch. 18.2 - Prob. 2LOCh. 18.3 - Describe the findings of the Human Genome Project.Ch. 18.3 - Prob. 2LOCh. 18.3 - Prob. 3LOCh. 18.4 - Prob. 1LOCh. 18.4 - Prob. 2LO
Ch. 18.4 - Prob. 3LOCh. 18.5 - Prob. 1LOCh. 18.5 - Prob. 2LOCh. 18.5 - Prob. 3LOCh. 18.6 - Prob. 1LOCh. 18 - Prob. 1DACh. 18 - If the human genome contains approximately 3...Ch. 18 - Prob. 1IQCh. 18 - Prob. 2IQCh. 18 - Prob. 3IQCh. 18 - Prob. 4IQCh. 18 - Prob. 5IQCh. 18 - Prob. 6IQCh. 18 - A genetic map provides a. the sequence of the DNA...Ch. 18 - Prob. 2UCh. 18 - Approximately how many genes are there in the...Ch. 18 - An open reading frame (ORF) is distinguished by...Ch. 18 - What is a BLAST search? a. A mechanism for...Ch. 18 - Prob. 6UCh. 18 - Prob. 7UCh. 18 - Prob. 8UCh. 18 - Prob. 1ACh. 18 - Prob. 2ACh. 18 - Prob. 3ACh. 18 - Prob. 4ACh. 18 - What information can be obtained from a DNA...Ch. 18 - Prob. 6ACh. 18 - Prob. 7ACh. 18 - You are in the early stages of a genome-sequencing...Ch. 18 - Genomic research can be used to determine if an...
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Molecular Biology Please help with question. Thank you in advance. Discuss, compare and contrast the structure of promoters inprokaryotes and eukaryotes.arrow_forwardMolecular Biology Please help with question. Thank you You are studying the expression of the lac operon. You have isolated mutants as described below. In the absence of glucose, explain/describe what would happen, for each mutant, to the expression of the lac operon when you add lactose AND what would happen when the bacteria has used up all of the lactose (if the mutant is able to use lactose).1. Mutations in the lac repressor gene that would prevent the binding of lactose2. Mutations in the lac repressor gene that would prevent release of lactose once lactose hadbound3. Normally the lac repressor gene is located next to (a few hundred base pairs) and upstreamfrom the lac operon. Mutations in the lac repressor gene that move the lac repressor gene 100,000base pairs downstream.4. Mutations in the lac operator that would prevent binding of lac repressorarrow_forwardYou have returned to college to become a phylogeneticist. One of the first things you wish to do is determine how mammals, birds, and reptiles are related. Like any good scientist, you need to consider all available data objectively and without a preconceived “correct” answer. In pursuit of that, you should produce a phylogenetic tree based only on morphological features that show birds and mammals are more closely related. You will then produce a totally different tree, also using morphological features, that shows birds and reptiles are more closely related. Do not forget to include all three groups in both your trees. Based solely off the trees you produce, which relationship would you consider the more likely and why? Once you have answered that question, provide a brief summary of the “modern” understanding of the relationship between these three groups.arrow_forward
- true or false, the reason geckos can walk on walls is hydrogen bonding between their foot pads and the moisture on the wall.arrow_forwardBiology laboratory problem Please help. thank you You have 20 ul of DNA solution and 6X DNA loading buffer solution. You have to mix your DNA solution and DNA loading buffer before load DNA in an agarose gel. The concentration of the DNA loading buffer must be 1X in the DNA and DNA-loading buffer mixture after you mix them. For that, I will add _____ ul of 6X loading buffer to the 20 ul DNA solution.arrow_forwardBiology lab problem To make 20 ul of 5 mM MgCl2 solution using 50 mM MgCl2 stock solution and distilled water, I will mix ________ ul of 50 mM MgCl2 solution and ________ ul of distilled water. Please help . Thank youarrow_forward
- Biology Please help. Thank you. Biology laboratory question You need 50 ml of 1% (w/v) agarose gel. Agarose is a powder. How would you make it? You can ignore the volume of agarose powder. Don't forget the unit.TBE buffer is used to make an agarose gel, not distilled water. I will add _______ of agarose powder into 50 ml of distilled water (final 50 ml).arrow_forwardAn urgent care center experienced the average patient admissions shown in the Table below during the weeks from the first week of December through the second week of April. Week Average Daily Admissions 1-Dec 11 2-Dec 14 3-Dec 17 4-Dec 15 1-Jan 12 2-Jan 11 3-Jan 9 4-Jan 9 1-Feb 12 2-Feb 8 3-Feb 13 4-Feb 11 1-Mar 15 2-Mar 17 3-Mar 14 4-Mar 19 5-Mar 13 1-Apr 17 2-Apr 13 Forecast admissions for the periods from the first week of December through the second week of April. Compare the forecast admissions to the actual admissions; What do you conclude?arrow_forwardAnalyze the effectiveness of the a drug treatment program based on the needs of 18-65 year olds who are in need of treatment by critically describing 4 things in the program is doing effectively and 4 things the program needs some improvement.arrow_forward
- I have the first half finished... just need the bottom half.arrow_forward13. Practice Calculations: 3 colonies were suspended in the following dilution series and then a viable plate count and microscope count was performed. Calculate IDF's, TDF's and then calculate the CFU/mL in each tube by both methods. Finally calculate the cells in 1 colony by both methods. Show all of your calculations in the space provided on the following pages. 3 colonies 56 cells 10 μL 10 μL 100 μL 500 με m OS A B D 5.0 mL 990 με 990 με 900 με 500 μL EN 2 100 με 100 μL 118 colonies 12 coloniesarrow_forwardDescribe and give a specific example of how successionary stage is related to species diversity?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning

Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning

Genome Annotation, Sequence Conventions and Reading Frames; Author: Loren Launen;https://www.youtube.com/watch?v=MWvYgGyqVys;License: Standard Youtube License