
Concept explainers
To estimate: The date when the most recent common ancestor of all three species lived using the given genetic data.
Introduction: A common ancestor of different species is an evolutionary link between the different species. Different organisms have evolved by differentiation and divergence from the common ancestor, which is their origin. The degree of differentiation or the time that it takes for an organism to evolve from a common ancestor is measured by the genetic distance.
Explanation of Solution
Pictorial representation: Fig.1 represents the phylogenetic tree of the common ancestry.
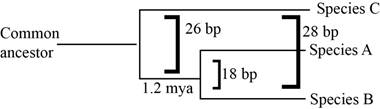
Fig.1: Phylogenetic tree
A species and B species are closely related, but species C is more distant to A and B. The phylogenetic tree shows that the common ancestor of A and B were seen 1.2 million years ago. As species C is even more distant, the common ancestor of all three was seen much before 1.2 million years ago. Since it took 1.2 million years to lead to a difference of 18 base pairs, time when the common ancestor on A, B, and C was seen, can be estimated to be 1.2 to 2 million years before the common ancestor of A and B.
To describe: The assumptions made to get the estimated date.
Introduction: A common ancestor of different species is an evolutionary link between the different species. Different organisms have evolved by differentiation and divergence from the common ancestor, which is their origin. The degree of differentiation or the time that it takes for an organism to evolve from a common ancestor is measured by the genetic distance.
Explanation of Solution
Species C is more distant to species A and B, hence their common ancestor is expected to present much earlier than the common ancestor of A and B. Species C is estimated to have lived 1.2 to 2 million years before the common ancestor of A and B.
To derive this, several assumptions are made such as follows:
- It is assumed that
speciation is a long and gradual process which occurs over a period of time. - Phylogenetic trees can be used to study diversification.
- It is assumed that the three species A, B, and C have some similarities and hence have a common ancestor, which serves as a point of origin for diversification of species.
Want to see more full solutions like this?
Chapter 7 Solutions
Evolution
- What is behavioral adaptarrow_forward22. Which of the following mutant proteins is expected to have a dominant negative effect when over- expressed in normal cells? a. mutant PI3-kinase that lacks the SH2 domain but retains the kinase function b. mutant Grb2 protein that cannot bind to RTK c. mutant RTK that lacks the extracellular domain d. mutant PDK that has the PH domain but lost the kinase function e. all of the abovearrow_forwardWhat is the label ?arrow_forward
- Can you described the image? Can you explain the question as well their answer and how to get to an answer to an problem like this?arrow_forwardglg 112 mid unit assignment Identifying melting processesarrow_forwardGive only the mode of inheritance consistent with all three pedigrees and only two reasons that support this, nothing more, (it shouldn't take too long)arrow_forward
- Oarrow_forwardDescribe the principle of homeostasis.arrow_forwardExplain how the hormones of the glands listed below travel around the body to target organs and tissues : Pituitary gland Hypothalamus Thyroid Parathyroid Adrenal Pineal Pancreas(islets of langerhans) Gonads (testes and ovaries) Placentaarrow_forward
 Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning





