
Concept explainers
To explain: The reason why the probability of death as a function of the body mass index is a U-shaped curve.
Concept introduction: The body mass index is the measure of the body fat based on the height and weight that applies to adult men and women. A body mass index between 18.5 and 25 is ideal. The body mass index between 25 and 30 is classified as overweight. The person with a BMI more than 30 is called obese. The BMI below 18 is classified as underweight person.
</SUMMARY-INTRODUCTION>
Pictorial representation: Fig.1 represents the “U” shaped curve of probability of death as a function of the body mass index.
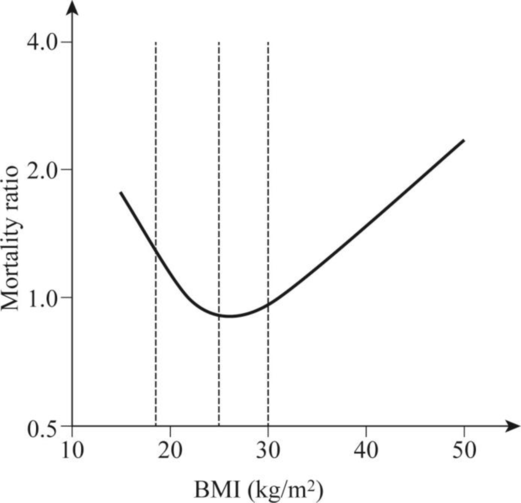
Fig. 1: “U” shaped curve of probability of death as a function of the body mass index.
Explanation:
The relationship between the all-cause mortality and body mass index in man is a U-shaped curve. The body mass index categorizes the people in overweight, normal weight, and underweight. The severely underweight individuals are at the risk of dying from the malnutrition-related causes. The individuals having 40% overweight have more chances of premature death when compared to a person with average weight. This is because obesity is linked to several serious medical conditions including diabetes and atherosclerosis. The individuals who belong to severely underweight categories and severely overweight categories are more likely to die from various reasons than the individuals who belong to normal category.
Want to see the full answer?
Check out a sample textbook solution
Chapter 22 Solutions
FUNDAMENTALS OF BIOCHEMISTRY - LL FD
- In a cell free preparation of beta-lactamase, penicillin is hydrolyzed in a D2O enriched assay. After one round of catalysis, where would you anticipate finding Deuterium? please help thank youarrow_forwardTo map the active site of -lactamase, the enzyme was hydrolyzed with trypsin to yield a hexapeptide (P1) with the following amino acids. Glu, Lys, Leu, Phe, Met, and Ser. Treatment of P1 with phenyl isothiocyanate yielded a PTH derivative of phenylalanine and a peptide (P2). Treatment of P1 with cyanogenbromide gave an acidic tetrapeptide (P3) and a dipeptide (P4).Treatment of P2 with 1-fluoro-2,4-dinitrobenzene, followed by complete hydrolysis, yields N-2,4-dinitrophenyl-Glu. P1, P2, and P3 contain the active site serine. question: the b-lactamase hydrolyzes the lactam-ring in antibiotics like penicillin. Describe the mechanism, of hydrolysis, insuring to include the involvement of S, D, and K in the reaction sequence. Please help!arrow_forwardThree of these amino acids participate in the proteolytic hydrolysis of polypeptides. Show the charge-relay network generated by the serine proteases and identify the nucleophilic species that initiates the hydrolysis. please help!arrow_forward
- You have isolated a protein and determined that the native molecular weight of the holoenzyme is 160 kD using size exclusion chromatography. Analysis of this protein using SDS-PAGE revealed 2 bands, one at 100 kD and one at 30 kD. 1. Describe the architecture of the polypeptide component of this enzyme. 2. The enzyme was found to be 0.829% NAD (by weight). What further can be said regarding the architecture? can you please help me with question number 2arrow_forwardTo map the active site of -lactamase, the enzyme was hydrolyzed with trypsin to yield a hexapeptide (P1) with the following amino acids. Glu, Lys, Leu, Phe, Met, and Ser. Treatment of P1 with phenyl isothiocyanate yielded a PTH derivative of phenylalanine and a peptide (P2). Treatment of P1 with cyanogenbromide gave an acidic tetrapeptide (P3) and a dipeptide (P4).Treatment of P2 with 1-fluoro-2,4-dinitrobenzene, followed by complete hydrolysis, yields N-2,4-dinitrophenyl-Glu. P1, P2, and P3 contain the active site serine. Question: although S, K, and D are involved in the catalysis, the E in this hexapeptide does not participate in the hydrolysis of the b-lactam ring. Why is that?arrow_forwardTo map the active site of beta-lactamase, the enzyme was hydrolyzed with trypsin to yield a hexapeptide (P1) with the following amino acids. Glu, Lys, Leu, Phe, Met, and Ser. a) Using the experimental results described below deduce the primary sequence of the active site hexapeptide. Treatment of P1 with phenyl isothiocyanate yielded a PTH derivative of phenylalanine and a peptide (P2). Treatment of P1 with cyanogenbromide gave an acidic tetrapeptide (P3) and a dipeptide (P4).Treatment of P2 with 1-fluoro-2,4-dinitrobenzene, followed by complete hydrolysis, yields N-2,4-dinitrophenyl-Glu. P1, P2, and P3 contain the active site serine. please help!arrow_forward
- The beta-lactamase hydrolyzes the lactam-ring in penicillin. Describe the mechanism of hydrolysis, insuring to include the involvement of S, D, & K in the reaction sequence. Please helparrow_forwardTo map the active site of beta-lactamase, the enzyme was hydrolyzed with trypsin to yield a hexapeptide (P1) with the following amino acids. Glu, Lys, Leu, Phe, Met, and Ser. Treatment of P1 with phenyl isothiocyanate yielded a PTH derivative of phenylalanine and a peptide (P2). Treatment of P1 with cyanogenbromide gave an acidic tetrapeptide (P3) and a dipeptide (P4).Treatment of P2 with 1-fluoro-2,4-dinitrobenzene, followed by complete hydrolysis, yields N-2,4-dinitrophenyl-Glu. P1, P2, and P3 contain the active site serine. Why doesn't D in this hexapeptide not participate in the hydrolysis of the beta-lactam ring even though S, K, and D are involved in the catalyst?arrow_forwardTo map the active site of -lactamase, the enzyme was hydrolyzed with trypsin to yield a hexapeptide (P1) with the following amino acids. Glu, Lys, Leu, Phe, Met, and Ser. Treatment of P1 with phenyl isothiocyanate yielded a PTH derivative of phenylalanine and a peptide (P2). Treatment of P1 with cyanogenbromide gave an acidic tetrapeptide (P3) and a dipeptide (P4).Treatment of P2 with 1-fluoro-2,4-dinitrobenzene, followed by complete hydrolysis, yields N-2,4-dinitrophenyl-Glu. P1, P2, and P3 contain the active site serine. Using the experimental results described above derive the primary sequence of the active site hexapeptide. Please help!arrow_forward
- Which type of enzyme catalyses the following reaction? oxidoreductase, transferase, hydrolase, lyase, isomerase, or ligase.arrow_forward+NH+ CO₂ +P H₂N + ATP H₂N NH₂ +ADParrow_forwardWhich type of enzyme catalyses the following reaction? oxidoreductase, transferase, hydrolase, lyase, isomerase, or ligase.arrow_forward
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON





