
Concept explainers
Review Figures 8.12 and 8.13. In cells, the primers for DNA synthesis are short strands of RNA, so each newly-synthesized strand of DNA has a segment of RNA al its 5′ end. As replication proceeds, DNA polymerases remove these RNA segments and fill in the resulting gaps with DNA. However, the gaps at the very 5′ ends of the new strands cannot be filled in with DNA. Why not?
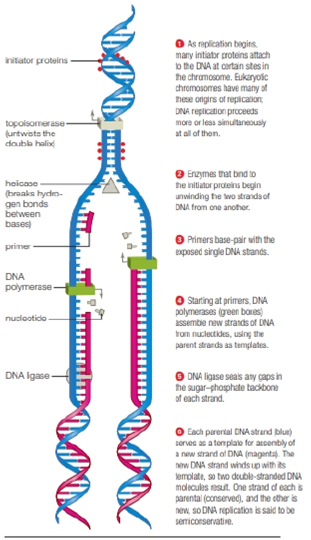
FIGURE 8.12 DNA replication.
Green arrows show the direction of synthesis for each strand. The Y-shaped structure where the DNA molecule is being unwound is called a replication fork.
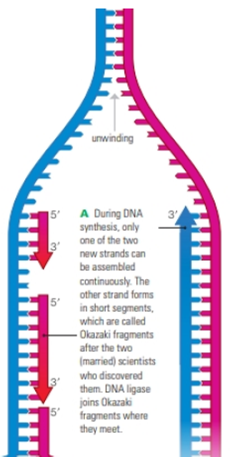
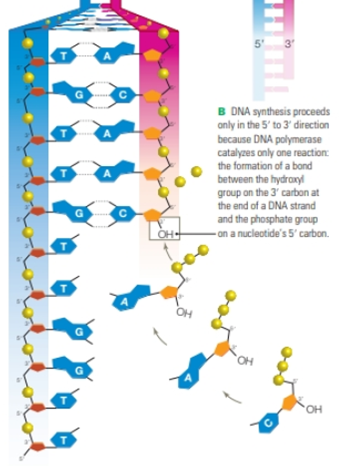
FIGURE 8.13 Discontinuous synthesis of DNA.
This close-up of a replication fork shows that only one of the two new DNA strands is assembled continuously. The other is assembled in short segments.
Trending nowThis is a popular solution!

Chapter 8 Solutions
Biology: The Unity and Diversity of Life (Looseleaf)
Additional Science Textbook Solutions
Biological Science (6th Edition)
College Physics: A Strategic Approach (3rd Edition)
Fundamentals of Physics Extended
EBK INTRODUCTION TO CHEMISTRY
Laboratory Manual For Human Anatomy & Physiology
- examples of synamtomorphy.arrow_forwardE. Bar Graph Use the same technique to upload the completed image. We will use a different type of graph to derive additional information from the CO2 data (Fig A1.6.2) 1. Calculate the average rate of increase in COz concentration per year for the time intervals 1959-1969, 1969- 1979, etc. and write the results in the spaces provided. The value for 1959-1969 is provided for you as an example. 2. Plot the results as a bar graph. The 1959-1969 is plotted for you. 3. Choose the graph that looks the most like yours A) E BAR GRAPH We will use a different type of graph to derive additional information from the CU, data (rig. nive). Average Yearly Rate of Observatory, Hawall interval Rate of increase per year 1959-1969 0.9 1969-1979 1979-1989 1989-1999 1999-2009 Figure A1.6.2 1999-2009 *- mrame -11- -n4 P2 جية 1989-1999 1979-1989 1969-1979 1959-1969 This bar drawn for you as an example 1.0 CO, Average Increase/Year (ppmv) B) E BAR GRAPH We will use a different type of graph to derive…arrow_forwardUse the relationships you just described to compute the values needed to fill in the blanks in the table in Fig A1.4.1 depth (a) 1.0 cml 0.7 cml cm| base dimensions (b, c)| 1.0 cm| 1.0 cm| 1.0 cm 1.0 cm| 1.0 cm| 1.0 cm volume (V) 1.0_cm' cm'| cm'| density (p) 1.0 g/cm'| 1.0 g/cm 1.0 g/cm' mass (m)| 0.3 g Column 1: depth at 1.0 cm volume mass Column 2: depth at 0.7 cm volume mass Column 3: unknown depth depth volumearrow_forward
- San Andreas Transform Boundary Plate Motion The geologic map below of southern California shows the position of the famous San Andreas Fault, a transform plate boundary between the North American Plate (east side) and the Pacific Plate (west side). The relative motion between the plates is indicated by the half arrows along the transform plate boundary (i.e., the Pacific Plate is moving to the northwest relative to the North American Plate). Note the two bodies of Oligocene volcanic rocks (labeled Ov) on the map in the previous page located along either side of the San Andreas Fault. These rocks are about 23.5 million years old and were once one body of rock. They have been separated by displacement along the fault. 21. Based on the offset of these volcanic rocks, what is the average annual rate of relative plate motion in cm/yr? SAF lab 2.jpg Group of answer choices 0.67 cm/yr 2 cm/yr 6.7 cm/yr 1.5 cm/yr CALIFORNIA Berkeley San Francisco K Os Q San Andreas Fault Ov…arrow_forwardThese are NOT part of any graded assignment. Are there other examples of synapomorphy. What is it called when the traits retained are similar to ancestors?arrow_forwardPlease hand draw everying. Thank you! Draw a gram positive bacterial cell below. Your cell should have the following parts, labeled: A coccus shape A capsule The gram positive cell wall should have the peptidoglycan labeled, as well as its component parts (NAM, NAG, and teichoic acid) A cell membrane Fimbriae A nucleoid Ribosomes Inclusionsarrow_forward
- Draw a gram negative bacterial cell below. Your cell should have the following parts, labeled: A bacillus shape Fimbriae Amphitrichous flagella 2 membranes (outer and inner) The outer membrane should have lipopolysaccharide (LPS) with lipid A and O antigens Periplasmic space The thin peptidoglycan cell wall between the 2 membranes A nucleoid Ribosomes Inclusionsarrow_forwardBacterial species Cell wall type Example: S. mitis Gram positive S. epidermidis H. pylori M. bovis S. marcescens Shape and arrangement Coccus, streptococcus Drawing 0000000arrow_forwardDraw a gram positive bacterial cell below. Your cell should have the following parts, labeled: A coccus shape A capsule The gram positive cell wall should have the peptidoglycan labeled, as well as its component parts (NAM, NAG, and teichoic acid) A cell membrane Fimbriae A nucleoid Ribosomes Inclusionsarrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning





