
Biochemistry: Concepts and Connections
1st Edition
ISBN: 9780321839923
Author: Dean R. Appling, Spencer J. Anthony-Cahill, Christopher K. Mathews
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Textbook Question
Chapter 4, Problem 8P
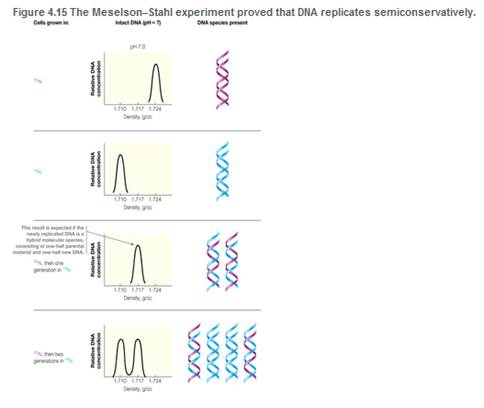
Refer to Figure 4.15, which presents the Meselson-Stahi experiment. DNA molecules can be denatured by high pH, as well as by heat. Suppose that the CsCl gradient centrifugations were run at pH 12, conditions under which DNA strands separate. Sketch the gradient profiles expected for each of the four samples depicted in the figure.
Expert Solution & Answer
Trending nowThis is a popular solution!

Students have asked these similar questions
Why the 2nd choice is correct?
a) What are the differences between the Direct & Indirect Immunofluorescence Assays? (0.5 mark)
b) What are the advantages of the Indirect Immunofluorescence Assays? (0.5 mark)
c) A Super-Resolution Imaging Technique was developed in 2018 using imidazole, a His-tag ligand conjugated with a fluorophore to report the presence of a recombinant His-tag protein target, (Sci Rep, 2018, 8:5507). How does this technique improve the image quality? (2 marks)
a) What are the differences between the Direct & Indirect Immunofluorescence Assays?
b) What are the advantages of the Indirect Immunofluorescence Assays?
c) A Super-Resolution Imaging Technique was developed in 2018 using imidazole, a His-tag ligand conjugated with a fluorophore to report the presence of a recombinant His-tag protein target, (Sci Rep, 2018, 8:5507). How does this technique improve the image quality?
Chapter 4 Solutions
Biochemistry: Concepts and Connections
Ch. 4 - Prob. 1PCh. 4 - What is the difference between a nucleoside...Ch. 4 - pppApCpCpupApGpApu-OH a. Using the straight-chain...Ch. 4 - Shown is a representation of a molecule being...Ch. 4 - Base analysis of DNA from maize (com) shows it to...Ch. 4 - Using the pKa data in Table 4.1 and the...Ch. 4 - For some DNAs, it is possible to separate the two...Ch. 4 - Refer to Figure 4.15, which presents the...Ch. 4 - Suppose that you centrifuged a transfer RNA...Ch. 4 - Predict the structure of a cruciform that could be...
Ch. 4 - DNA from a newly discovered virus was purified,...Ch. 4 - Would you expect Neurospora crassa DNA to have a...Ch. 4 - A circular double-stranded DNA molecule contains...Ch. 4 - The gel electrophoresis pattern in Figure 4.23 was...Ch. 4 - 15. DNA polymerase requires both a template, to be...Ch. 4 - Prob. 16P
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biochemistry and related others by exploring similar questions and additional content below.Similar questions
- Calculate the number of ATP produced from oxidation of 1 molecule of glucosearrow_forwardExample 1: 1. Suppose an enzyme (MW = 5,000 g/mole) has a concentration of 0.05 mg/L. If the kcat is 1 x 10 s, what is the theoretical maximum reaction velocity for the enzyme? A) 1050 µM/s. B) 100 µM/s. C) 150 μM/s. D) 105 μM/s.arrow_forwardIn 1956, E. P. Kennedy and S. B. Weiss published their study of membrane lipid phosphatidylcholine (lecithin) synthesis in rat liver. Their hypothesis was that phosphocholine joined with some cellular component to yield lecithin. In an earlier experiment, incubating 32 P-labeled phosphocholine at physiological temperature (37 °C) with broken cells from rat liver yielded labeled lecithin. This became their assay for the enzymes involved in lecithin synthesis. Determine the optimal pH for this enzyme and characterize the enzyme activity at different pH values. -O-P-O-CH2-CH₁₂-N(CH3)3 Phosphocholine H₂C-O-C-R HC-O-C-R2 + + + Cell fraction + ? HC-O-P-O-CH₁₂-CH₂-N(CH), O Phosphatidylcholine The researchers then centrifuged the broken cell preparation to separate the membranes from the soluble proteins. They tested three preparations: whole extract, membranes, and soluble proteins. Table 1 summarizes the results. Table 1: Cell fraction requirement for incorporation of 32p-phosphocholine into…arrow_forward
- Researchers isolated an unknown substance, X, from rabbit muscle. They determined its structure from the following observations and experiments. (a) Qualitative analysis showed that X was composed entirely of C, H, and O. A weighed sample of X was completely oxidized and the H2O and CO2 produced were measured. This quantitative analysis revealed that X contained 40.00% C, 6.71% H, and 53.29% O by weight. (b) The molecular mass of X, as determined by mass spectrometry, was 90.00 atomic mass units (u). (c) Infrared spectroscopy showed that X contained one double bond. (d) X dissolved readily in water, and the solution demonstrated optical activity when tested in a polarimeter. (e) The aqueous solution of X is acidic. What is the empirical formula of X?arrow_forwardShow work. don't give Ai generated solution....give correct solutionarrow_forwardBiochemistry What is the process of "transamination" in either the muscles or the liver, that involves keto acid or glutamic acid? Please explain how the steps work. Thank you!arrow_forward
- Biochemistry Please help. Thank you What is the importance of glutamic acid in the metabolism of nitrogen from amino acids? (we know therole; it’s used to remove the nitrogen from amino acids so that the remaining carbon skeleton can bebroken down by the “usual” pathways, but what is the important, unique role that only glutamicacid/glutamate can do?)arrow_forwardBiochemistry Please help. Thank you When carbamyl phosphate is joined to L-ornathine, where does the energy for the reaction come from?arrow_forwardBiochemistry Question Please help. Thank you What is the function of glutamate dehydrogenase?arrow_forward
- Biochemistry Question Please help. Thank you How and why does a high protein diet affect the enzymes of the urea cycle?arrow_forwardBiochemistry What is the importance of the glucose-alanine cycle?arrow_forwardBiochemistry Assuming 2.5 molecules of ATP per oxidation of NADH/(H+) and 1.5molecules of ATP per oxidation of FADH2, how many ATP are produced per molecule of pyruvate? Please help. Thank youarrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College

DNA Use In Forensic Science; Author: DeBacco University;https://www.youtube.com/watch?v=2YIG3lUP-74;License: Standard YouTube License, CC-BY
Analysing forensic evidence | The Laboratory; Author: Wellcome Collection;https://www.youtube.com/watch?v=68Y-OamcTJ8;License: Standard YouTube License, CC-BY