
PRESCOTT'S MICROBIOLOGY
11th Edition
ISBN: 2818440045677
Author: WILLEY
Publisher: MCG
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 18.3, Problem 2CC
Retrieve, Infer, Apply
Examine figure 18.8. How might metagenomics be used to isolate genes encoding a potentially new antibiotic?
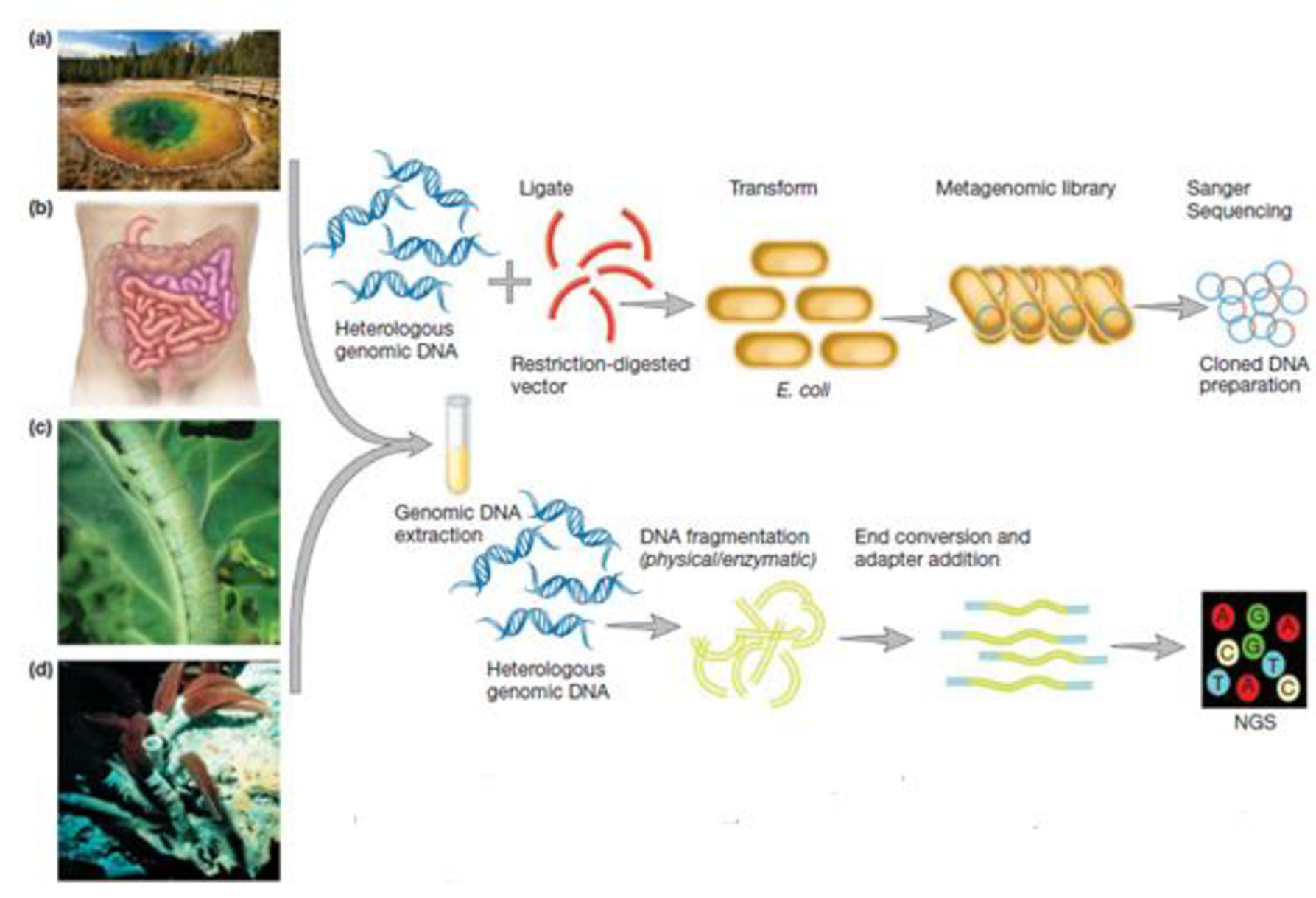
Figure 18.8 Metagenomic analysis. DNA has been extracted directly from environments such as (a) bacterial mats at Yellowstone National Park, (b) the human colon, (c) cabbage white butterfly larvae, and (d) tube worms from hydrothermal vents. The DNA can be used to construct a metagenomic library and analyzed by Sanger sequencing (top); however, NGS is more likely to be used (bottom), thereby avoiding the need for genomic library construction.
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Question 4
1 pts
Which of the following would be most helpful for demonstrating alternative splicing for a
new organism?
○ its proteome and its transcriptome
only its transcriptome
only its genome
its proteome and its genome
If the metabolic scenario stated with 100 mM of a sucrose solution, how much ATP would be made then during fermentation?
What is agricu
Chapter 18 Solutions
PRESCOTT'S MICROBIOLOGY
Ch. 18.1 - MICRO INQUIRY What is the function of the 3-OH...Ch. 18.1 - MICRO INQUIRY Why is it important that identical...Ch. 18.2 - MICRO INQUIRY Which step (or steps) in this...Ch. 18.2 - Retrieve, Infer, Apply Why is the Sanger technique...Ch. 18.2 - Retrieve, Infer, Apply Explain the difference...Ch. 18.2 - Retrieve, Infer, Apply Why does reversible chain...Ch. 18.2 - Prob. 4CCCh. 18.2 - Retrieve, Infer, Apply Suggest a medical and an...Ch. 18.3 - Retrieve, Infer, Apply NGS techniques are...Ch. 18.3 - Retrieve, Infer, Apply Examine figure 18.8. How...
Ch. 18.4 - Prob. 1MICh. 18.4 - Prob. 1CCCh. 18.4 - Prob. 2CCCh. 18.4 - Prob. 3CCCh. 18.5 - Figure 18.12 Metabolic Pathways and Transport...Ch. 18.5 - Prob. 2MICh. 18.5 - Prob. 3MICh. 18.5 - Prob. 1CCCh. 18.5 - Retrieve, Infer, Apply How might the following...Ch. 18.5 - Retrieve, Infer, Apply Compare and contrast...Ch. 18.5 - Retrieve, Infer, Apply Why does two-dimensional...Ch. 18.5 - Retrieve, Infer, Apply What is the difference...Ch. 18.5 - Retrieve, Infer, Apply Describe a ChIP-Seq...Ch. 18.7 - Prob. 1MICh. 18.7 - Retrieve, Infer, Apply Cite an infectious disease...Ch. 18.7 - Prob. 2CCCh. 18.7 - Prob. 3CCCh. 18 - Prob. 1RCCh. 18 - Prob. 2RCCh. 18 - Prob. 3RCCh. 18 - Prob. 4RCCh. 18 - Prob. 5RCCh. 18 - Prob. 1ALCh. 18 - Prob. 2ALCh. 18 - You are developing a new vaccine for a pathogen....Ch. 18 - Prob. 4AL
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- When using the concept of "a calorie in is equal to a calorie out" how important is the quality of the calories?arrow_forwardWhat did the Cre-lox system used in the Kikuchi et al. 2010 heart regeneration experiment allow researchers to investigate? What was the purpose of the cmlc2 promoter? What is CreER and why was it used in this experiment? If constitutively active Cre was driven by the cmlc2 promoter, rather than an inducible CreER system, what color would you expect new cardiomyocytes in the regenerated area to be no matter what? Why?arrow_forwardWhat kind of organ size regulation is occurring when you graft multiple organs into a mouse and the graft weight stays the same?arrow_forward
- What is the concept "calories consumed must equal calories burned" in regrads to nutrition?arrow_forwardYou intend to insert patched dominant negative DNA into the left half of the neural tube of a chick. 1) Which side of the neural tube would you put the positive electrode to ensure that the DNA ends up on the left side? 2) What would be the internal (within the embryo) control for this experiment? 3) How can you be sure that the electroporation method itself is not impacting the embryo? 4) What would you do to ensure that the electroporation is working? How can you tell?arrow_forwardDescribe a method to document the diffusion path and gradient of Sonic Hedgehog through the chicken embryo. If modifying the protein, what is one thing you have to consider in regards to maintaining the protein’s function?arrow_forward
- The following table is from Kumar et. al. Highly Selective Dopamine D3 Receptor (DR) Antagonists and Partial Agonists Based on Eticlopride and the D3R Crystal Structure: New Leads for Opioid Dependence Treatment. J. Med Chem 2016.arrow_forwardThe following figure is from Caterina et al. The capsaicin receptor: a heat activated ion channel in the pain pathway. Nature, 1997. Black boxes indicate capsaicin, white circles indicate resinferatoxin. You are a chef in a fancy new science-themed restaurant. You have a recipe that calls for 1 teaspoon of resinferatoxin, but you feel uncomfortable serving foods with "toxins" in them. How much capsaicin could you substitute instead?arrow_forwardWhat protein is necessary for packaging acetylcholine into synaptic vesicles?arrow_forward
- 1. Match each vocabulary term to its best descriptor A. affinity B. efficacy C. inert D. mimic E. how drugs move through body F. how drugs bind Kd Bmax Agonist Antagonist Pharmacokinetics Pharmacodynamicsarrow_forward50 mg dose of a drug is given orally to a patient. The bioavailability of the drug is 0.2. What is the volume of distribution of the drug if the plasma concentration is 1 mg/L? Be sure to provide units.arrow_forwardDetermine Kd and Bmax from the following Scatchard plot. Make sure to include units.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning


Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biology (MindTap Course List)
Biology
ISBN:9781337392938
Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. Berg
Publisher:Cengage Learning

Case Studies In Health Information Management
Biology
ISBN:9781337676908
Author:SCHNERING
Publisher:Cengage

Molecular Techniques: Basic Concepts; Author: Dr. A's Clinical Lab Videos;https://www.youtube.com/watch?v=7HFHZy8h6z0;License: Standard Youtube License