
Campbell Biology 11th Edition - Valuepack
11th Edition
ISBN: 9780134833545
Author: Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Jane B. Reece Neil A. Campbell Lisa A. Urry
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 18, Problem 11TYU
draw it The diagram below shows five genes, including their enhancers, from the genome of a certain species. Imagine that yellow, blue, green, black, red, and purple activator proteins exist that can bind to the appropriately color-coded control elements in the enhancers of these genes.
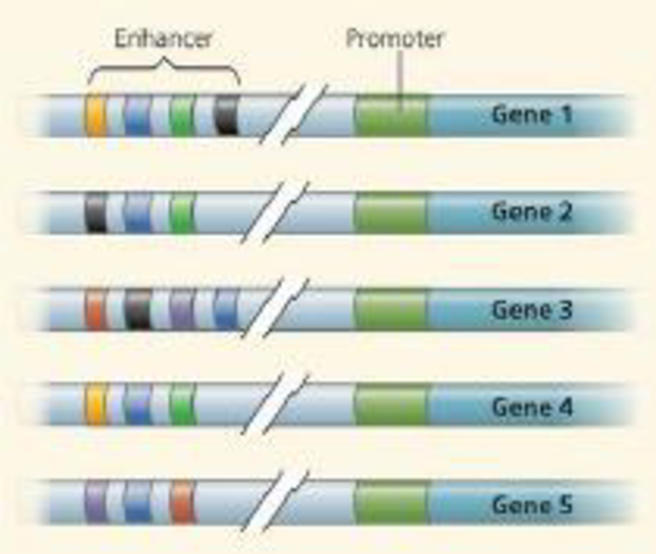
(a) Draw an X above enhancer elements (of all the genes) that would have activators bound in a cell in which only gene 5 is transcribed. Identify which colored activators would be present.
- (b) Draw a dot above all enhancer elements that would have activators bound in a cell in which the green, blue, and yellow activators are present. Identify which gene(s) would be transcribed.
- (c) Imagine that genes 1,2, and 4 code for nerve-specific proteins, and genes 3 and 5 are skin specific. Identify which activators would have to be present in each cell type to ensure transcription of the appropriate genes.
Expert Solution & Answer
Trending nowThis is a popular solution!

Students have asked these similar questions
O Macmillan Learning
You have 0.100 M solutions of acetic acid (pKa = 4.76) and sodium acetate. If you wanted to prepare 1.00 L of 0.100 M acetate
buffer of pH 4.00, how many milliliters of acetic acid and sodium acetate would you add?
acetic acid:
mL
sodium acetate:
mL
How does the cost of food affect the nutritional choices people make?
Biopharmaceutics and Pharmacokinetics:Two-Compartment Model Zero-Order Absorption Questions
SHOW ALL WORK, including equation used, variables used and each step to your solution, report your regression lines and axes names (with units if appropriate) :Calculate a-q
a) B1,
b) B2,
c) hybrid rate constant (1)
d) hybrid rate constant (2)
e) t1/2,dist
f) t1/2,elim
g) k10
h) k12
i) k21
j) initial concentration (C0)
k) central compartment volume (V1)
l) steady-state volume (Vss)
m) clearance (CL) AUC (0→10 min) using trapezoidal rule
n) AUC (20→30 min) using trapezoidal rule
o) AUCtail (AUC360→∞)
p) total AUC (using short cut method)
q) volume from AUC (VAUC)
Chapter 18 Solutions
Campbell Biology 11th Edition - Valuepack
Ch. 18.1 - How does binding of the trp corepressor to the trp...Ch. 18.1 - Describe the binding of RNA Polymerase,...Ch. 18.1 - WHAT IF? A certain mutation in E. coli changes...Ch. 18.2 - In general, what are the effects of histone...Ch. 18.2 - MAKE CONNECTIONS Speculate about whether the same...Ch. 18.2 - Compare the roles of general and specific...Ch. 18.2 - Once mRNA encoding a particular protein reaches...Ch. 18.2 - WHAT IF? Suppose you compared the nucleotide...Ch. 18.3 - Compare miRNAs and siRNAs, including their...Ch. 18.3 - WH AT IF? Suppose the mRNA being degraded in...
Ch. 18.3 - MAKE CONNECTIONS Inactivation of one of the X...Ch. 18.4 - MAKE CONNECTIONS As you learned in Chapter 12,...Ch. 18.4 - MAKE CONNECTIONS Explain how the signaling...Ch. 18.4 - How do fruit fly maternal effect genes determine...Ch. 18.4 - Prob. 4CCCh. 18.5 - Prob. 1CCCh. 18.5 - Under what circumstances is cancer considered to...Ch. 18.5 - MAKE CONNECTIONS The p53 protein can activate...Ch. 18 - Compare and contrast the roles of a corepressor...Ch. 18 - Describe what must happen in a cell for a gene...Ch. 18 - Why are miRNAs called noncoding RNAs? Explsin how...Ch. 18 - Describe the two main processes that cause...Ch. 18 - Compare the usual functions of proteins encoded by...Ch. 18 - If a particular operon encodes enzymes for making...Ch. 18 - Muscle cells differ from nerve cells mainly...Ch. 18 - The functioning of enhancers is an example of (A)...Ch. 18 - Cell differentiation always involves (A)...Ch. 18 - Which of the following is an example of...Ch. 18 - What would occur if the repressor of an inducible...Ch. 18 - Absence of bicoid in mRNA from a Drosophila egg...Ch. 18 - Which of the following statements about the DNA in...Ch. 18 - Within a cell, the amount of protein made using a...Ch. 18 - Prob. 10TYUCh. 18 - draw it The diagram below shows five genes,...Ch. 18 - Prob. 12TYUCh. 18 - Prob. 13TYUCh. 18 - SCIENCE. TECHNOLOGY, AND SOCIETY Trace amounts of...Ch. 18 - WRITE ABOUT A THEME: INTERACTIONS In a Short essay...Ch. 18 - SYNTHESIZE YOUR KNOWLEDGE The flashlight fish has...
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- What are some external influences that keep people from making healthy eating decisions?arrow_forwardWhat type of structure(s) would you expect to see in peripheral membrane proteins? (mark all that apply) A. Amphipathic alpha helix (one side is hydrophilic and one side is hydrophobic) B. A hydrophobic beta barrel C. A hydrophobic alpha helix D. A chemical group attached to the protein that can anchor it to the membranearrow_forwardTemporal flexibility (the ability to change over time) of actin structures within a cell is maintained by… A. The growth/shrinkage cycle B. Periodic catastrophe C. GTP hydrolysis D. Treadmilling E. None of the abovearrow_forward
- During in vitro polymerization of actin and microtubule filaments from their subunits, what causes the initial delay in filament growth? A.Nucleation B.Reaching homeostasis C.Nucleotide exchange D.ATP or GTP hydrolysis E.Treadmillingarrow_forwardYou expect to find which of the following in the Microtubule Organizing Center (MTOC)...(mark all that apply) A. Gamma tubulin B. XMAP215 C. Centrioles D. Kinesin-13arrow_forwardThe actin-nucleating protein formin has flexible “arms” containing binding sites that help recruit subunits in order to enhance microfilament polymerization. What protein binds these sites? A.Thymosin B.Profilin C.Cofilin D.Actin E.Tropomodulinarrow_forward
- While investigating an unidentified motor protein, you discover that it has two heads that bind to actin. Based on this information, you could confidently determine that it is NOT... (mark all that apply) A. A myosin I motor B. A dynein motor C. A myosin VI motor D. A kinesin motorarrow_forwardYou isolate the plasma membrane of cells and find that . . . A. it contains regions with different lipid compositions B. it has different lipid types on the outer and cytosolic leaflets of the membrane C. neither are possible D. A and B both occurarrow_forwardYou are studying the mobility of a transmembrane protein that contains extracellular domains, one transmembrane domain, and a large cytosolic domain. Under normal conditions, this protein is confined to a particular region of the membrane due to the cortical actin cytoskeletal network. Which of the following changes is most likely to increase mobility of this protein beyond the normal restricted region of the membrane? A. Increased temperature B. Protease cleavage of the extracellular domain of the protein C. Binding to a free-floating extracellular ligand, such as a hormone D. Protease cleavage of the cytosolic domain of the protein E. Aggregation of the protein with other transmembrane proteinsarrow_forward
- Topic: Benthic invertebrates as an indicator species for climate change, mapping changes in ecosystems (Historical Analysis & GIS) What objects or events has the team chosen to analyze? How does your team wish to delineate the domain or scale in which these objects or events operate? How does that limited domain facilitate a more feasible research project? What is your understanding of their relationships to other objects and events? Are you excluding other things from consideration which may influence the phenomena you seek to understand? Examples of such exclusions might include certain air-born pollutants; a general class of water bodies near Ottawa, or measurements recorded at other months of the year; interview participants from other organizations that are involved in the development of your central topic or issue. In what ways do your research questions follow as the most appropriate and/or most practical questions (given the circumstances) to pursue to better understand…arrow_forwardThe Esp gene encodes a protein that alters the structure of the insulin receptor on osteoblasts and interferes with the binding of insulin to the receptor. A researcher created a group of osteoblasts with an Esp mutation that prevented the production of a functional Esp product (mutant). The researcher then exposed the mutant strain and a normal strain that expresses Esp to glucose and compared the levels of insulin in the blood near the osteoblasts (Figure 2). Which of the following claims is most consistent with the data shown in Figure 2 ? A Esp expression is necessary to prevent the overproduction of insulin. B Esp protein does not regulate blood-sarrow_forwardPredict the per capita rate of change (r) for a population of ruil trees in the presence of the novel symbiont when the soil moisture is 29%. The formula I am given is y= -0.00012x^2 + 0.0088x -0.1372. Do I use this formula and plug in 29 for each x variable?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning

Biology (MindTap Course List)
Biology
ISBN:9781337392938
Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. Berg
Publisher:Cengage Learning

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning


Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning
An Introduction to the Human Genome | HMX Genetics; Author: Harvard University;https://www.youtube.com/watch?v=jEJp7B6u_dY;License: Standard Youtube License