
Concept explainers
Answers to all problems are at the end οΓthis book. Detailed solutions are available in the Student Solutions Manual. Study Guide, and Problems Book.
Calculation of Rate Enhancement from Energies of Activation
The relationships between the free energy terms defined in the solution to Problem 4 earlier are shown in the following figure.
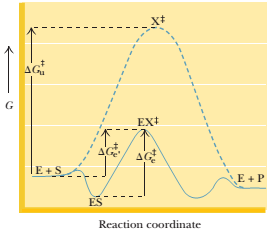
If the energy of the ES complex is 10 kJ/mol lower than the energy of E + S, the value of
Want to see the full answer?
Check out a sample textbook solution
Chapter 14 Solutions
Study Guide with Student Solutions Manual and Problems Book for Garrett/Grisham's Biochemistry, 6th
- Br Mg, ether 1. HCHO (formaldehyde) 2. H+, H₂O PCC 1. NH3, HCN ? (pyridinium chlorochromate) 2. H2O, HCI 11. Which one of the following compounds is the major organic product of the series of reactions shown above? Ph. Ph. OH NH2₂ A Ph. Ή NH2 B OH Ph Η Ph OH NH2 NH2₂ NH₂ C D Earrow_forwardB A 6. Which ONE of the labeled bonds in the tripeptide on the right is a peptide bond: H₂N N 'N' OH C H A, B, C, D or E? HN E OHarrow_forwardQuestions 8-9 are 0.4 points each. The next two questions relate to the peptide whose structure is shown here. To answer these questions, you should look at a table of H2N/.. amino acid structures. You don't have to memorize the structures of the amino acids. IZ 8. What is the N-terminal amino acid of this peptide? A) proline B) aspartic acid C) threonine 9. What is the C-terminal amino acid of this peptide? A) proline B) aspartic acid C) threonine N OH D) valine E) leucine D) valine E) leucine NH "OH OHarrow_forward
- 7. What is the correct name of the following tripeptide? A) Ile-Met-Ser B) Leu-Cys-Thr C) Val-Cys-Ser D) Ser-Cys-Leu E) Leu-Cys-Ser H₂N!!!!! N H ΖΙ .SH SF H IN OH OHarrow_forwardPlease draw out the following metabolic pathways: (Metabolic Map) Mitochondrion: TCA Cycle & GNG, Electron Transport, ATP Synthase, Lipolysis, Shuttle Systems Cytoplasm: Glycolysis & GNG, PPP (Pentose Phosphate Pathway), Glycogen, Lipogenesis, Transporters and Amino Acids Control: Cori/ Glc-Ala cycles, Insulin/Glucagon Reg, Local/Long Distance Regulation, Pools Used Correctlyarrow_forwardPlease help provide me an insight of what to draw for the following metabolic pathways: (Metabolic Map) Mitochondrion: TCA Cycle & GNG, Electron Transport, ATP Synthase, Lipolysis, Shuttle Systems Cytoplasm: Glycolysis & GNG, PPP (Pentose Phosphate Pathway), Glycogen, Lipogenesis, Transporters and Amino Acids Control: Cori/ Glc-Ala cycles, Insulin/Glucagon Reg, Local/Long Distance Regulation, Pools Used Correctlyarrow_forward
- f. The genetic code is given below, along with a short strand of template DNA. Write the protein segment that would form from this DNA. 5'-A-T-G-G-C-T-A-G-G-T-A-A-C-C-T-G-C-A-T-T-A-G-3' Table 4.5 The genetic code First Position Second Position (5' end) U C A G Third Position (3' end) Phe Ser Tyr Cys U Phe Ser Tyr Cys Leu Ser Stop Stop Leu Ser Stop Trp UCAG Leu Pro His Arg His Arg C Leu Pro Gln Arg Pro Leu Gin Arg Pro Leu Ser Asn Thr lle Ser Asn Thr lle Arg A Thr Lys UCAG UCAC G lle Arg Thr Lys Met Gly Asp Ala Val Gly Asp Ala Val Gly G Glu Ala UCAC Val Gly Glu Ala Val Note: This table identifies the amino acid encoded by each triplet. For example, the codon 5'-AUG-3' on mRNA specifies methionine, whereas CAU specifies histidine. UAA, UAG, and UGA are termination signals. AUG is part of the initiation signal, in addition to coding for internal methionine residues. Table 4.5 Biochemistry, Seventh Edition 2012 W. H. Freeman and Company B eviation: does it play abbreviation:arrow_forwardAnswer all of the questions please draw structures for major productarrow_forwardfor glycolysis and the citric acid cycle below, show where ATP, NADH and FADH are used or formed. Show on the diagram the points where at least three other metabolic pathways intersect with these two.arrow_forward
 BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
