
Concept explainers
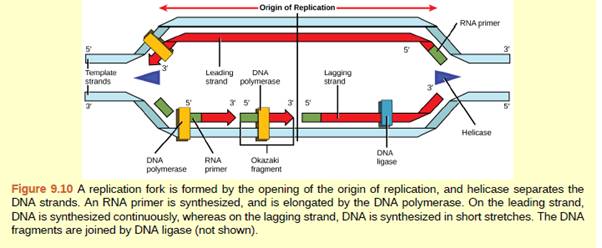
Figure 9.10 You isolate a cell strain in which the joining together of Okazaki fragments is impaired and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is most likely to be mutated?
To write:
The enzyme which is most likely mutated when joining of Okazaki fragments is impaired in the replication.
Introduction:
DNA replication is the process in which the copy of DNA is made. This is done in 3 steps in a eukaryotic cell. They are initiation, elongation, and termination. It is a semiconservative type of replication. Enzymes involved in this process are helicase, DNA polymerase, and DNA ligase.
Explanation of Solution
DNA replication is started at the origin of replication where a Y shaped fork known as replication is formed when both strands are separated by helicase enzyme. Both the strands of DNA act as a template. One strand is known as a leading strand at which replication is continuous and other strand is known as a lagging strand at which replication occurs in fragments. This is because DNA polymerase can synthesize DNA only in 5' to 3' direction. These fragments are known as Okazaki fragments. These fragments at the lagging strands are then sealed by the enzyme DNA ligase. The joining of Okazaki fragments is impaired when the DNA ligase is not able to join the strands. This can occur due to the mutation in enzyme DNA ligase.
When joining of DNA fragments on the lagging strand is impaired in DNA replication, the mutation is most likely to occur in DNA ligase as it is an important enzyme which joins the strands.
Want to see more full solutions like this?
Chapter 9 Solutions
Concepts of Biology
Additional Science Textbook Solutions
College Physics: A Strategic Approach (3rd Edition)
Campbell Biology (11th Edition)
Genetic Analysis: An Integrated Approach (3rd Edition)
Biology: Life on Earth (11th Edition)
Campbell Essential Biology (7th Edition)
Human Anatomy & Physiology (2nd Edition)
- 13. Practice Calculations: 3 colonies were suspended in the following dilution series and then a viable plate count and microscope count was performed. Calculate IDF's, TDF's and then calculate the CFU/mL in each tube by both methods. Finally calculate the cells in 1 colony by both methods. Show all of your calculations in the space provided on the following pages. 3 colonies 56 cells 10 μL 10 μL 100 μL 500 με m OS A B D 5.0 mL 990 με 990 με 900 με 500 μL EN 2 100 με 100 μL 118 colonies 12 coloniesarrow_forwardDescribe and give a specific example of how successionary stage is related to species diversity?arrow_forwardExplain down bellow what happens to the cell in pictures not in words: Decreased pH in mitochondria Increased ATP Decreased pH in cytosol Increased hydrolysis Decreasing glycogen and triglycerides Increased MAP kinase activity Poor ion transport → For each one:→ What normally happens?→ What is wrong now?→ How does it mess up the cell?arrow_forward
- 1.) Community Diversity: The brown and orange line represent two different plant communities. a. Which color represents the community with a higher species richness? b. Which color represents the community with a higher species evenness? Relative abundance 0.1 0.04 0.001 2 4 6 8 10 12 14 16 18 20 22 24 Rank abundance c. What is the maximum value of the Simpson's diversity index (remember, Simpson's index is D = p², Simpson's diversity index is 1-D)? d. If the Simpson's diversity index equals 1, what does that mean about the number of species and their relative abundance within community being assessed?arrow_forward1.) Community Diversity: The brown and orange line represent two different plant communities. a. Which color represents the community with a higher species richness? b. Which color represents the community with a higher species evenness? Relative abundance 0.1 0.04 0.001 2 4 6 8 10 12 14 16 18 20 22 24 Rank abundance c. What is the maximum value of the Simpson's diversity index (remember, Simpson's index is D = p², Simpson's diversity index is 1-D)? d. If the Simpson's diversity index equals 1, what does that mean about the number of species and their relative abundance within community being assessed?arrow_forwardwhat measures can a mother to take to improve the produce of her to milk to her newborn baby ?arrow_forward
- 1. Color the line that represents all ancestors of the Eastern white pine tree green (but only the ancestral line NOT shared with other organisms) 2. Oncle the last common ancestor of the Colorado blue spruce tree and Eastern white pine tree. 3. Put a box around the last common ancestor of the sugar maple tree and the dogwood tree. 4. Put a triangle around the last common ancestor of the red pine tree and the american holly bush. 5. Color the line that represents all ancestors of the Ponderosa pine tree red (including all shared ancestors). 6. Color the line that represents all ancestors of the American elm tree blue (including all shared ancestors). 7 Color the line that represents all ancestors of the Sabal palm tree purple (including all shared ancestors) 8. Using a yellow highlighter or colored pencil, circle the clade that includes all pine trees. 9. Using a orange highlighter or colored pencil, circle the clade that includes all gymnosperms 10. Can you tell…arrow_forwardYou have been hired as a public relations specialist to give invertebrates a good name. After all, they are much more than just creepy crawly bugs! Your first task though is to convince yourself that is true. The best way to do that is to start close to home. Find something in your house that is a product obtained directly from an invertebrate or only due to an invertebrate’s actions. Describe the product, its function and utility, as well as any human manufactured alternatives. Be sure to highlight the advantages of obtaining this directly from nature. Keep in mind, a product can be something you use, wear, eat, or enjoy for its visual appeal.arrow_forwardUse the following tree diagram to answer Questions #8-10. 8) Which of the following two animals are the most closely related based on the tree to the left? a) Pig and camel b) Hippo and pig c) Deer and cow 9) CIRCLE on the tree diagram where the common ancestor between a hippo and a cow is. 10) Put a SQUARE on the tree diagram where the common ancestor between a pig and a peccary is.arrow_forward
- Explain: Healthy Cell Function Overview→ Briefly describe how a healthy cell usually works: metabolism (ATP production), pH balance, glycogen storage, ion transport, enzymes, etc. Gene Mutation and Genetics Part→ Focus on the autosomal recessive mutation and explain: How gene mutation affects the cell. How autosomal inheritance works. Compare the normal and mutated gene sequences simply. → Talk about possible consequences of a faulty hydrolytic enzyme.arrow_forwardCan you fill out those termsarrow_forwardExplain down bellow what happens to the cell: Decreased pH in mitochondria Increased ATP Decreased pH in cytosol Increased hydrolysis Decreasing glycogen and triglycerides Increased MAP kinase activity Poor ion transport → For each one:→ What normally happens?→ What is wrong now?→ How does it mess up the cell?arrow_forward
 Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning





