
Concept explainers
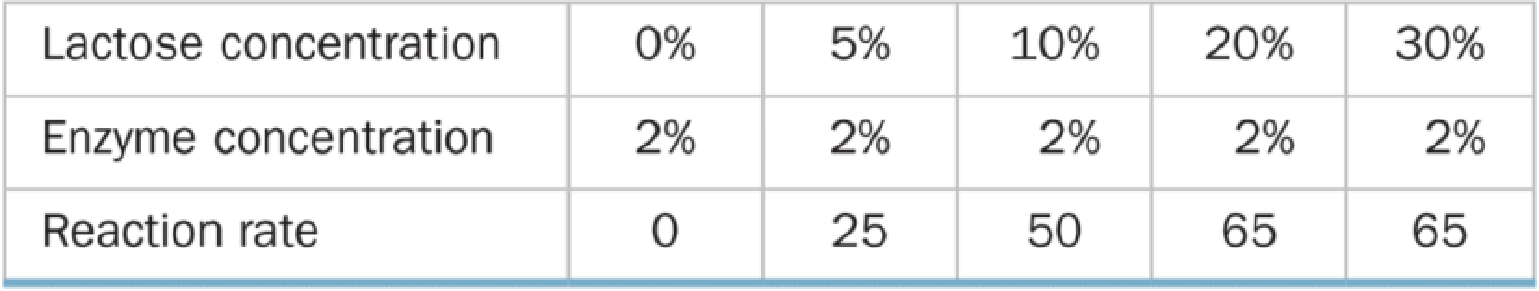
A biologist performed two series of experiments on lactase, the enzyme that hydrolyzes lactose to glucose and galactose. First, she made up 10% lactose solutions containing different concentrations of enzyme and measured the rate at which galactose was produced (grams of galactose per minute). Results of these experiments are shown in Table A below. In the second series of experiments (Table B), she prepared 2% enzyme solutions containing different concentrations of lactose and again measured the rate of galactose production.
Table A Reaction Rate and Enzyme Concentration
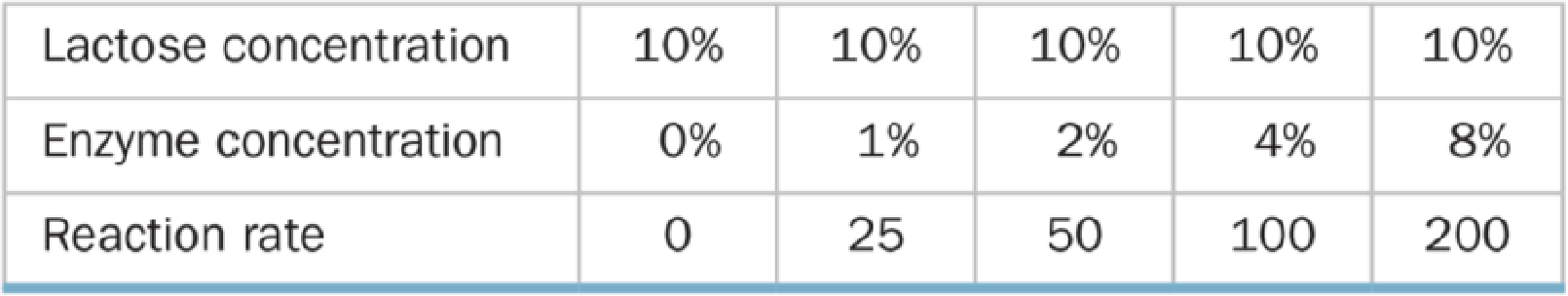
Table B Reaction Rate and Substrate Concentration
a. Graph and explain the relationship between the reaction rate and the enzyme concentration.
b. Graph and explain the relationship between the reaction rate and the substrate concentration. How and why did the results of the two experiments differ?
Trending nowThis is a popular solution!

Chapter 5 Solutions
Campbell Biology: Concepts & Connections (8th Edition)
- A negligence action was brought by a mother against a hospital on behalf of her minor daughter. It alleged that when the mother was 13 years of age, the hospital negligently transfused her with Rh-positive blood. The mother's Rh-negative blood was incompatible with and sensitized by the Rh-positive blood. The mother discovered her condition 8 years later during a routine blood screening ordered by her healthcare provider in the course of prenatal care. The resulting sensitization of the mother's blood allegedly caused damage to the fetus, resulting in physical defects and premature birth. Did a patient relationship with the transfusing hospital exist?arrow_forward18. Watch this short youtube video about SARS CoV-2 replication. SARS-CoV-2 Life Cycle (Summer 2020) - YouTube.19. What is the name of the receptor that SARS CoV-2 uses to enter cells? Which human cells express this receptor? 20. Name a few of the proteins that the SARS CoV-2 mRNA codes for. 21. What is the role of the golgi apparatus related to SARS CoV-2arrow_forwardState the five functions of Globular Proteins, and give an example of a protein for each function.arrow_forward
- Diagram of check cell under low power and high powerarrow_forwarda couple in which the father has the a blood type and the mother has the o blood type produce an offspring with the o blood type, how does this happen? how could two functionally O parents produce an offspring that has the a blood type?arrow_forwardWhat is the opening indicated by the pointer? (leaf x.s.) stomate guard cell lenticel intercellular space none of thesearrow_forward
- Identify the indicated tissue? (stem x.s.) parenchyma collenchyma sclerenchyma ○ xylem ○ phloem none of thesearrow_forwardWhere did this structure originate from? (Salix branch root) epidermis cortex endodermis pericycle vascular cylinderarrow_forwardIdentify the indicated tissue. (Tilia stem x.s.) parenchyma collenchyma sclerenchyma xylem phloem none of thesearrow_forward
- Identify the indicated structure. (Cucurbita stem l.s.) pit lenticel stomate tendril none of thesearrow_forwardIdentify the specific cell? (Zebrina leaf peel) vessel element sieve element companion cell tracheid guard cell subsidiary cell none of thesearrow_forwardWhat type of cells flank the opening on either side? (leaf x.s.) vessel elements sieve elements companion cells tracheids guard cells none of thesearrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College- Case Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
 Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage LearningEssentials Health Info Management Principles/Prac...Health & NutritionISBN:9780357191651Author:BowiePublisher:Cengage
Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage LearningEssentials Health Info Management Principles/Prac...Health & NutritionISBN:9780357191651Author:BowiePublisher:Cengage





