
GENERAL,ORGANIC, & BIOLOGICAL CHEM-ACCES
4th Edition
ISBN: 9781265982959
Author: SMITH
Publisher: MCG
expand_more
expand_more
format_list_bulleted
Concept explainers
Question
Chapter 22, Problem 48P
Interpretation Introduction
Interpretation:
Each of the letters (A-F) should be labeled in the below diagram as
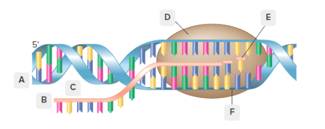
Concept Introduction:
Transcription is the process which copies the information stored in a DNA strand as a nucleotide sequence into a messenger RNA (mRNA) strand as an RNA nucleotide sequence.
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Calculate the pH and the pOH of each of the following solutions at 25 °C for which the substances ionize completely:
(a) 0.200 M HCl
Calculate the pH and the pOH of each of the following solutions at 25 °C for which the substances ionize completely:
(a) 0.000259 M HClO4
What is the pH of a 1.0 L buffer made with 0.300 mol of HF (Ka = 6.8 × 10⁻⁴) and 0.200 mol of NaF to which 0.160 mol of NaOH were added?
Chapter 22 Solutions
GENERAL,ORGANIC, & BIOLOGICAL CHEM-ACCES
Ch. 22.1 - Prob. 22.1PPCh. 22.1 - Prob. 22.1PCh. 22.1 - -Fluorouracil is an anticancer drug that...Ch. 22.1 - Prob. 22.3PCh. 22.1 - Prob. 22.4PCh. 22.1 - Label each statement about the compound...Ch. 22.1 - Draw the structure of each nucleotide: (a) UMP;...Ch. 22.1 - Give the name that corresponds to each...Ch. 22.2 - Draw the structure of a dinucleotide formed by...Ch. 22.2 - Draw the structure of each polynucleotide: (a) CU;...
Ch. 22.2 - Label each statement about the polynucleotide...Ch. 22.3 - Write the complementary strand for each of the...Ch. 22.4 - What is the sequence of a newly synthesized DNA...Ch. 22.6 - For each DNA segment: [1] What is the sequence of...Ch. 22.6 - Prob. 22.9PCh. 22.7 - What amino acid is coded for by each codon? GCC...Ch. 22.7 - What codons code for each amino acid? a. glycine...Ch. 22.7 - Drive the amino acid sequence that is coded for by...Ch. 22.7 - Write a possible mRNA sequence that codes for each...Ch. 22.7 - Considering the given sequence of nucleotides in...Ch. 22.8 - Prob. 22.14PCh. 22.8 - Prob. 22.8PPCh. 22.8 - Prob. 22.9PPCh. 22.9 - Prob. 22.10PPCh. 22.9 - Prob. 22.15PCh. 22.10 - Prob. 22.16PCh. 22 - Label each statement as pertaining to DNA, RNA, or...Ch. 22 - Label each statement as pertaining to DNA, RNA, or...Ch. 22 - Prob. 19PCh. 22 - (a) Give the name of each compound shown as a...Ch. 22 - Prob. 21PCh. 22 - Prob. 22PCh. 22 - Prob. 23PCh. 22 - Prob. 24PCh. 22 - Prob. 25PCh. 22 - Draw the structure of each of the following: a...Ch. 22 - Prob. 27PCh. 22 - Prob. 28PCh. 22 - Prob. 29PCh. 22 - Prob. 30PCh. 22 - Prob. 31PCh. 22 - Draw the structures of the two possible...Ch. 22 - Prob. 33PCh. 22 - Draw the structure of each dinucleotide and...Ch. 22 - Draw the deoxyribonucleotide TGA. Label the 5 and...Ch. 22 - Draw the ribonucleotide CGU. Label the 5 and 3...Ch. 22 - Prob. 37PCh. 22 - Describe in detail the DNA double helix with...Ch. 22 - Write the sequence of the complementary strand of...Ch. 22 - Prob. 40PCh. 22 - Prob. 41PCh. 22 - Prob. 42PCh. 22 - Prob. 43PCh. 22 - Prob. 44PCh. 22 - Prob. 45PCh. 22 - Figure 22.4 snows the hydrogen-bonding...Ch. 22 - Prob. 47PCh. 22 - Prob. 48PCh. 22 - Prob. 49PCh. 22 - Prob. 50PCh. 22 - What mRNA is transcribed from each DNA sequence in...Ch. 22 - Prob. 52PCh. 22 - For each DNA segment: [1] What is the sequence of...Ch. 22 - Prob. 54PCh. 22 - For each codon, give its anticodon and the amino...Ch. 22 - For each codon, give its anticodon and the amino...Ch. 22 - Fill in the missing information in the schematic...Ch. 22 - Fill in the missing information in the schematic...Ch. 22 - Derive the amino acid sequence that is coded for...Ch. 22 - Derive the amino acid sequence that is coded for...Ch. 22 - Write a possible mRNA sequence that codes for each...Ch. 22 - Prob. 62PCh. 22 - Prob. 63PCh. 22 - Prob. 64PCh. 22 - Prob. 65PCh. 22 - Prob. 66PCh. 22 - Consider the following mRNA sequence:...Ch. 22 - Consider the following mRNA sequence: 5-ACC UUA...Ch. 22 - Consider the following sequence of DNA: 3-TTA...Ch. 22 - Consider the following sequence of DNA: 3-ATA...Ch. 22 - Prob. 71PCh. 22 - Prob. 72PCh. 22 - Prob. 73PCh. 22 - Prob. 74PCh. 22 - Prob. 75PCh. 22 - Prob. 76PCh. 22 - Prob. 77PCh. 22 - Prob. 78PCh. 22 - Prob. 79PCh. 22 - Prob. 80PCh. 22 - Prob. 81PCh. 22 - Prob. 82PCh. 22 - Fill in the base, codon, anticodon, or amino acid...Ch. 22 - Fill in the base, codon, anticodon, or amino acid...Ch. 22 - Fill in the base, codon, anticodon, or amino acid...Ch. 22 - Prob. 86PCh. 22 - Prob. 87PCh. 22 - Prob. 88PCh. 22 - Prob. 89PCh. 22 - Prob. 90PCh. 22 - Prob. 91CPCh. 22 - Give a possible nucleotide sequence in the...
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, chemistry and related others by exploring similar questions and additional content below.Similar questions
- Determine if the following salt is neutral, acidic or basic. If acidic or basic, write the appropriate equilibrium equation for the acid or base that exists when the salt is dissolved in aqueous solution. If neutral, simply write only NR. Be sure to include the proper phases for all species within the reaction. NaN₃arrow_forwardCan I please get help with this?arrow_forwardCan I please get help with this?arrow_forward
- Use the Henderson-Hasselbalch equation to calculate pH of a buffer containing 0.050M benzoic acidand 0.150M sodium benzoate. The Ka of benzoic acid is 6.5 x 10-5arrow_forwardA. Draw the structure of each of the following alcohols. Then draw and name the product you would expect to produce by the oxidation of each. a. 4-Methyl-2-heptanol b. 3,4-Dimethyl-1-pentanol c. 4-Ethyl-2-heptanol d. 5,7-Dichloro-3-heptanolarrow_forwardWhat is the pH of a 1.0 L buffer made with 0.300 mol of HF (Ka = 6.8 × 10⁻⁴) and 0.200 mol of NaF to which 0.160 mol of NaOH were added?arrow_forward
- Can I please get help with this.arrow_forwardDetermine if the following salt is neutral, acidic or basic. If acidic or basic, write the appropriate equilibrium equation for the acid or base that exists when the salt is dissolved in aqueous solution. If neutral, simply write only NR. Be sure to include the proper phases for all species within the reaction. N₂H₅ClO₄arrow_forwardPlease help me with identifying these.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Chemistry for Today: General, Organic, and Bioche...ChemistryISBN:9781305960060Author:Spencer L. Seager, Michael R. Slabaugh, Maren S. HansenPublisher:Cengage Learning
Chemistry for Today: General, Organic, and Bioche...ChemistryISBN:9781305960060Author:Spencer L. Seager, Michael R. Slabaugh, Maren S. HansenPublisher:Cengage Learning General, Organic, and Biological ChemistryChemistryISBN:9781285853918Author:H. Stephen StokerPublisher:Cengage Learning
General, Organic, and Biological ChemistryChemistryISBN:9781285853918Author:H. Stephen StokerPublisher:Cengage Learning Organic And Biological ChemistryChemistryISBN:9781305081079Author:STOKER, H. Stephen (howard Stephen)Publisher:Cengage Learning,
Organic And Biological ChemistryChemistryISBN:9781305081079Author:STOKER, H. Stephen (howard Stephen)Publisher:Cengage Learning,- Chemistry: Matter and ChangeChemistryISBN:9780078746376Author:Dinah Zike, Laurel Dingrando, Nicholas Hainen, Cheryl WistromPublisher:Glencoe/McGraw-Hill School Pub Co
 Introduction to General, Organic and BiochemistryChemistryISBN:9781285869759Author:Frederick A. Bettelheim, William H. Brown, Mary K. Campbell, Shawn O. Farrell, Omar TorresPublisher:Cengage Learning
Introduction to General, Organic and BiochemistryChemistryISBN:9781285869759Author:Frederick A. Bettelheim, William H. Brown, Mary K. Campbell, Shawn O. Farrell, Omar TorresPublisher:Cengage Learning Chemistry & Chemical ReactivityChemistryISBN:9781337399074Author:John C. Kotz, Paul M. Treichel, John Townsend, David TreichelPublisher:Cengage Learning
Chemistry & Chemical ReactivityChemistryISBN:9781337399074Author:John C. Kotz, Paul M. Treichel, John Townsend, David TreichelPublisher:Cengage Learning

Chemistry for Today: General, Organic, and Bioche...
Chemistry
ISBN:9781305960060
Author:Spencer L. Seager, Michael R. Slabaugh, Maren S. Hansen
Publisher:Cengage Learning

General, Organic, and Biological Chemistry
Chemistry
ISBN:9781285853918
Author:H. Stephen Stoker
Publisher:Cengage Learning

Organic And Biological Chemistry
Chemistry
ISBN:9781305081079
Author:STOKER, H. Stephen (howard Stephen)
Publisher:Cengage Learning,

Chemistry: Matter and Change
Chemistry
ISBN:9780078746376
Author:Dinah Zike, Laurel Dingrando, Nicholas Hainen, Cheryl Wistrom
Publisher:Glencoe/McGraw-Hill School Pub Co

Introduction to General, Organic and Biochemistry
Chemistry
ISBN:9781285869759
Author:Frederick A. Bettelheim, William H. Brown, Mary K. Campbell, Shawn O. Farrell, Omar Torres
Publisher:Cengage Learning

Chemistry & Chemical Reactivity
Chemistry
ISBN:9781337399074
Author:John C. Kotz, Paul M. Treichel, John Townsend, David Treichel
Publisher:Cengage Learning
Nucleic acids - DNA and RNA structure; Author: MEDSimplified;https://www.youtube.com/watch?v=0lZRAShqft0;License: Standard YouTube License, CC-BY