
Concept explainers
- a)
Interpretation: The number of unpaired electrons in the given complex ions to be predicted.
Concept Introduction:
Crystal field splitting: The splitting of d-orbitals in the presence of ligands is known as crystal field splitting.
Spectrochemical series: The list of ligands arranged in an ascending order of
Crystal field splitting: The energy gap between the splitting of d-orbitals of the metal ion in presence of ligands is known as the crystal field splitting
To Identify: The number of unpaired electrons in the given complex ions to be predicted.
- a)
Answer to Problem 22.32QP
The complex ion
Explanation of Solution
Interpret the complex ion
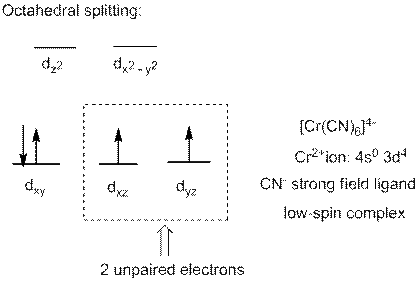
In complex ion
- b)
Interpretation: The number of unpaired electrons in the given complex ions to be predicted.
Concept Introduction:
Crystal field splitting: The splitting of d-orbitals in the presence of ligands is known as crystal field splitting.
Spectrochemical series: The list of ligands arranged in an ascending order of
Crystal field splitting: The energy gap between the splitting of d-orbitals of the metal ion in presence of ligands is known as the crystal field splitting
To Identify: The number of unpaired electrons in the given complex ions to be predicted.
- b)
Answer to Problem 22.32QP
The complex ion
Explanation of Solution
Interpret the complex ion
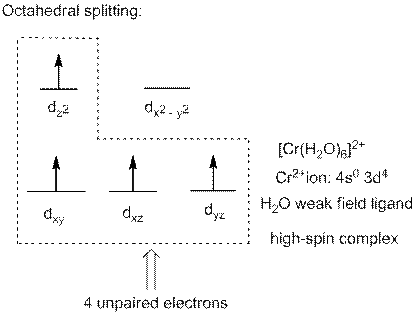
In complex ion
Want to see more full solutions like this?
Chapter 22 Solutions
Chemistry: Atoms First V1
- Predict the intermediate 1 and final product 2 of this organic reaction: NaOMe H+ + 1 2 H H work up You can draw 1 and 2 in any arrangement you like. Note: if either 1 or 2 consists of a pair of enantiomers, just draw one structure using line bonds instead of 3D (dash and wedge) bonds at the chiral center. Click and drag to start drawing a structure. X $ dmarrow_forwardPredict the major products of this organic reaction: 1. NaH (20°C) 2. CH3Br ? Some notes: • Draw only the major product, or products. You can draw them in any arrangement you like. • Be sure to use wedge and dash bonds where necessary, for example to distinguish between major products that are enantiomers. • If there are no products, just check the box under the drawing area. No reaction. Click and drag to start drawing a structure. G Crarrow_forwardPredict the major products of this organic reaction: 1. LDA (-78°C) ? 2. Br Some notes: • Draw only the major product, or products. You can draw them in any arrangement you like. . • Be sure to use wedge and dash bonds where necessary, for example to distinguish between major products that are enantiomers. • If there are no products, just check the box under the drawing area. No reaction. Click and drag to start drawing a structure. Xarrow_forward
- Please draw the structuresarrow_forwardDraw the missing intermediates 1 and 2, plus the final product 3, of this synthesis: 0 1. Eto 1. Eto- 1 2 2. MeBr 2. EtBr H3O+ A 3 You can draw the three structures in any arrangement you like. Explanation Check Click and drag to start drawing a structure.arrow_forwardDraw the missing intermediate 1 and final product 2 of this synthesis: 1. MeO- H3O+ 1 2 2. PrBr Δ You can draw the two structures in any arrangement you like. Click and drag to start drawing a structure.arrow_forward
- What is the differences between: Glyceride and phosphoglyceride Wax and Fat Soap and Fatty acid HDL and LDL cholesterol Phospho lipids and sphingosine What are the types of lipids? What are the main lipid components of membrane structures? How could lipids play important rules as signaling molecules and building units? The structure variety of lipids makes them to play significant rules in our body, conclude breifly on this statement.arrow_forwardWhat is the differences between DNA and RNA for the following: - structure - function - type What is the meaning of: - replication - transcription - translation show the base pair connection(hydrogen bond) in DNA and RNAarrow_forwardWhat is the IP for a amino acid- give an example what are the types of amino acids What are the structures of proteins The N-Terminal analysis by the Edman method shows saralasin contains sarcosine at the N-terminus. Partial hydrolysis of saralasin with dilute hydrochloric acid yields the following fragments: Try-Val-His Sar-Arg-Val His-Pro-Ala Val- Tyr- Val Arg-Val-Tyr What is the structure of saralasin?arrow_forward
- What is the IP for a amino acid- give an example what are the types of amino acids What are the structures of proteins The N-Terminal analysis by the Edman method shows saralasin contains sarcosine at the N-terminus. Partial hydrolysis of saralasin with dilute hydrochloric acid yields the following fragments: Try-Val-His Sar-Arg-Val His-Pro-Ala Val- Tyr- Val Arg-Val-Tyr What is the structure of saralasin?arrow_forward> aw the missing intermediates 1 and 2, plus the final product 3, of this synthesis: 1. Eto 1. EtO¯ H3O+ 1 2 2. PrBr 2. PrBr Δ You can draw the three structures in any arrangement you like. 3 Click and drag to start drawing a structure. Explanation Check 2025 McGraw Hill LLC. All Rights Reserved. Terms of Use Privacarrow_forwardThere are various factors that affect an equilibrium. Give 3 of these factors and explain using examples andequations how an equilibrium is affected by these factors. Please remember that this is a communication question so that you are communicating your understanding of the factors that affect and equilibrium.arrow_forward
 ChemistryChemistryISBN:9781305957404Author:Steven S. Zumdahl, Susan A. Zumdahl, Donald J. DeCostePublisher:Cengage Learning
ChemistryChemistryISBN:9781305957404Author:Steven S. Zumdahl, Susan A. Zumdahl, Donald J. DeCostePublisher:Cengage Learning Chemistry: An Atoms First ApproachChemistryISBN:9781305079243Author:Steven S. Zumdahl, Susan A. ZumdahlPublisher:Cengage Learning
Chemistry: An Atoms First ApproachChemistryISBN:9781305079243Author:Steven S. Zumdahl, Susan A. ZumdahlPublisher:Cengage Learning
 Chemistry: The Molecular ScienceChemistryISBN:9781285199047Author:John W. Moore, Conrad L. StanitskiPublisher:Cengage Learning
Chemistry: The Molecular ScienceChemistryISBN:9781285199047Author:John W. Moore, Conrad L. StanitskiPublisher:Cengage Learning Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning
Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning Chemistry: Principles and PracticeChemistryISBN:9780534420123Author:Daniel L. Reger, Scott R. Goode, David W. Ball, Edward MercerPublisher:Cengage Learning
Chemistry: Principles and PracticeChemistryISBN:9780534420123Author:Daniel L. Reger, Scott R. Goode, David W. Ball, Edward MercerPublisher:Cengage Learning





