
Concept explainers
(a)
To describe: The chemical transformation which is required to convert glutamate to (2S)-4-amino-2-hydroxybutyrate (AHBA).
Introduction:
Glutamate comes under the category of non-essential amino acid and it contains two carboxyl groups. It is an acidic amino acid which is important part of the neurotransmission process. AHBA (4-amino-2-bydoxy butyrate) it is main component of many antibiotics.
(a)
Explanation of Solution
Pictorial representation: Fig.1 shows formation of AHBA from glutamate.
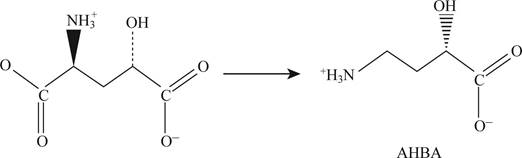
Fig.1: Formation of AHBA from glutamate.
Explanation:
AHBA (4-amino-2-bydoxy butyrate) is formed from the glutamate by the process of decarboxylation. When there is a removal of α-carboxyl group from the glutamate and addition of one –OH group to the γ-carbon then AHBA is formed which is given in Fig. 1.
(b)
To determine: How Btrl can act as an acyl carrier protein with a Coenzyme A prosthetic group.
Introduction:
Coenzyme A is important in the synthesis and oxidation of the fatty acids and it also help in the citric acid cycle during the oxidation of the pyruvate.
(b)
Explanation of Solution
Explanation:
Btrl and acyl carrier proteins have similarities in terms of their sequences. When this compound incubated in CoA conditions it increases its molecular weight. It is because CoA molecule can bind to Btrl molecule when CoA is added to the serine residue (any one) with the replacement of the hydroxyl group with a 4’-phosphopantertheine group
The molecular weight of the CoA is 17;
And the molecular weight of 4’phosphopantetheine is 356;
Molecular mass of purified Btrl protein is 11,812.
So, the overall molecular weight of the Btrl which could act as an acyl carrier protein with a Coenzyme A would be as follows:
This calculated molecular weight is very much close to the observed molecular weight 12,153.
Conclusion:
Btrl can act as an acyl carrier protein with a Coenzyme A because the expected molecular weight is very close to the calculated value of molecular weight which is 12,153.
(c)
To determine: The structures which are consistent with the proposed structure of γ-glutamyl-S-Btrl.
Introduction:
Alpha carboxyl group is any organic molecule group that contains basic amino group and acidic carboxyl group which is very unique to each amino acid.
(c)
Explanation of Solution
Pictorial representation: Fig. 2 shows glutamyl-S-Btrl.
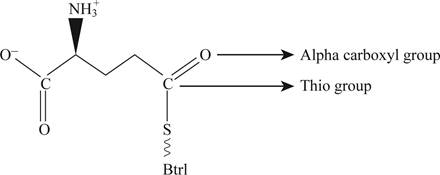
Fig.2: Glutamyl-S-Btrl
Explanation:
One more structure which is consistent is that a thio-ester could formed with the α-carboxyl group which is represented in the Fig. 2. Thio-ester is the compound which has
(d)
To determine: The chemical basis of the γ-glutamyl-S-Btrl can be correct because the alpha carboxyl group might be removed at some stage in the process.
Introduction:
Organic molecule group such as alpha carboxyl group that contains basic amino group and acidic carboxyl group and it is very unique to each amino acid.
(d)
Explanation of Solution
Explanation:
The process of removal of a carboxyl group from a compound is termed as decarboxylation. In many reactions, carboxyl group should be in a free state for an amino acid to be removed. It is very difficult to imagine that decarboxylation process can occur with a carboxyl group in its thio-ester form.
(e)
To determine: The most likely structure of the BtrK species
Introduction:
Decarboxylation is a
(e)
Explanation of Solution
Pictorial representation: Fig 3 shows most likely structure of the BtrK
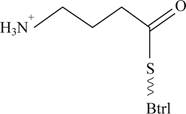
Fig 3: Most likely structure of the BtrK
Explanation:
The molecular species of Mr is 12,240 when the γ-glutymyl-S-Btrl is incubated with purified Btrl.
The molecular species of Mr is 12,281 of γ-glutymyl-S-Btrl.
Therefore,
Due to decarboxylation of the BtrK, carboxyl group would be deleted from the structure and only
(f)
To determine: The molecular structure of the species whose molecular mass is 12,370.
Introduction:
A pure substance is made from molecules with the average geometrical structure. The chemical formula and the structure of a molecule are the two important factors that determine.
(f)
Explanation of Solution
Pictorial representations: Fig 4 shows expected molecular structure of the species whose molecular mass is 12,370.
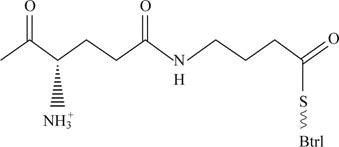
Fig 4: Expected molecular structure of the species whose molecular mass is 12,370.
Explanation:
The Molecular structure of the species whose molecular mass is 12,370 is represented in the figure 4. The molecular species of Mr is 12,240 when the γ-glutymyl-S-Btrl is incubated with purified Btrl. The Molecular structure of the species whose molecular mass is 12,370.
Therefore,
This molecular structure would have free amino group and alpha carboxyl group and thio group will remain in the new species.
(g)
To propose: The plausible pathway for the synthesis of AHBA with the help of enzymes that catalyze each step and any kind of cofactors and substrate required during the pathway.
Introduction:
In the plausible pathways different kind of enzyme and cofactor and substrate are used with the help of these factors a useful product can be synthesized.
(g)
Explanation of Solution
Pictorial representation: Fig.5 shows the plausible pathway for t.he synthesis of AHBA
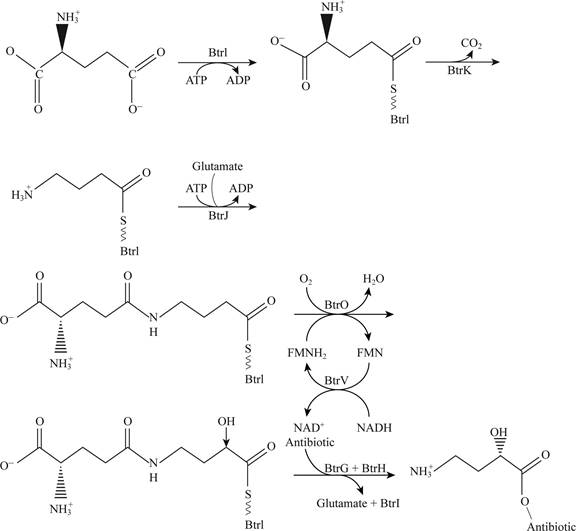
Fig.5: The plausible pathway for the synthesis of AHBA
Explanation:
The plausible pathway for the synthesis of AHBA and respective addition of antibiotics is represented in the Fig. 5. This pathway includes the involvement of co-factor such as ATP, NAD and glutamate amino acid.
Want to see more full solutions like this?
Chapter 22 Solutions
Lehninger Principles of Biochemistry
- By malonic or acetylacetic synthesis, synthesize 2-methylbutanoic acid (indicate the formulas of the compounds).arrow_forwardObtain 2-methylbutanoic acid by malonic or acetylacetic synthesis (indicate the formulas of the compounds involved).arrow_forwardEFFICIENTS SAMPLE READINGS CONCENTRATIONS Pigiadient) TOMATO SAUCE (REGULAR) TOMATO (REDUCED SALT) TOMATO SAUCE (REGULAR) TOMATO (REDUCED SALT) 58 6.274 3.898 301.7 151.2 14150 5.277 3.865 348.9 254.8 B 5.136 3.639 193.7 85.9 605 4.655 3.041 308.6 199.6 05 5.135 3.664 339.5 241.4 0139 4.676 3.662 160.6 87.6 90148 5.086 3.677 337.7 242.5 0092 6.348 3.775 464.7 186.4 PART3 5.081 3.908 223.5 155.8 5.558 3.861 370.5 257.1 4.922 3.66 326.6 242.9 4.752 3.641 327.5 253.3 50 5.018 3.815 336.1 256.0 84 4.959 3.605 317.9 216.6 38 4.96 3.652 203.8 108.7 $3 5.052 3.664 329.8 239.0 17 5.043 3.767 221.9 149.7 052 5.058 3.614 331.7 236.4 5.051 4.005 211.7 152.1 62 5.047 3.637 309.6 222.7 5.298 3.977 223.4 148.7 5.38 4.24 353.7 278.2 5 5.033 4.044 334.6 268.7 995 4.706 3.621 305.6 234.4 04 4.816 3.728 340.0 262.7 16 4.828 4.496 304.3 283.2 0.011 4.993 3.865 244.7 143.6 AVERAGE STDEV COUNT 95% CI Confidence Interval (mmol/L) [Na+] (mg/100 mL) 95% Na+ Confidence Interval (mg/100 mL)arrow_forward
- If we have two compounds: acetone (CH₃COCH₃) and acetic acid (CH₃COOH), applying heat to them produces an aldol condensation of the two compounds. If this is correct, draw the formula for the final product.arrow_forwardIf we have two compounds: acetone (CH3COCH3) and acetic acid (CH3COOH); if we apply heat (A), what product(s) are obtained?arrow_forwardQUESTION: Fill out the answers to the empty green boxes attached in the image. *Ensure you all incorporate all 27 values (per column)*arrow_forward
- You need to make a buffer by dissolving benzoic acid and sodium benzoate in water. What is the mass of benzoic acid that you would weigh out, in mg, to create 50 mL of a buffer at pH = 4.7 that will change pH no more than 0.10 units with the addition of 0.001 moles of acid or base? Enter just the answer without the units (mg) - just the number will do!arrow_forwardDraw the formula for 3-isopropylcyclopentane-1-carbonyl chloride.arrow_forwardQUESTION: Fill out the answers to the empty green boxes attached in the image. *Ensure you all incorporate all 27 values (per column)*arrow_forward
 ChemistryChemistryISBN:9781305957404Author:Steven S. Zumdahl, Susan A. Zumdahl, Donald J. DeCostePublisher:Cengage Learning
ChemistryChemistryISBN:9781305957404Author:Steven S. Zumdahl, Susan A. Zumdahl, Donald J. DeCostePublisher:Cengage Learning ChemistryChemistryISBN:9781259911156Author:Raymond Chang Dr., Jason Overby ProfessorPublisher:McGraw-Hill Education
ChemistryChemistryISBN:9781259911156Author:Raymond Chang Dr., Jason Overby ProfessorPublisher:McGraw-Hill Education Principles of Instrumental AnalysisChemistryISBN:9781305577213Author:Douglas A. Skoog, F. James Holler, Stanley R. CrouchPublisher:Cengage Learning
Principles of Instrumental AnalysisChemistryISBN:9781305577213Author:Douglas A. Skoog, F. James Holler, Stanley R. CrouchPublisher:Cengage Learning Organic ChemistryChemistryISBN:9780078021558Author:Janice Gorzynski Smith Dr.Publisher:McGraw-Hill Education
Organic ChemistryChemistryISBN:9780078021558Author:Janice Gorzynski Smith Dr.Publisher:McGraw-Hill Education Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning
Chemistry: Principles and ReactionsChemistryISBN:9781305079373Author:William L. Masterton, Cecile N. HurleyPublisher:Cengage Learning Elementary Principles of Chemical Processes, Bind...ChemistryISBN:9781118431221Author:Richard M. Felder, Ronald W. Rousseau, Lisa G. BullardPublisher:WILEY
Elementary Principles of Chemical Processes, Bind...ChemistryISBN:9781118431221Author:Richard M. Felder, Ronald W. Rousseau, Lisa G. BullardPublisher:WILEY





