
Concept explainers
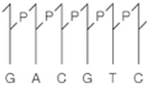
(a) Interpretation:
The other half of a restriction site should be drawn if the one-half is as follows:
Concept Introduction:
The restriction site also known as the restriction sequence is an enzyme that splits DNA into fragments at a recognized site inside a molecule. The restriction site's other part is given and the type II cleavage site with the blunt-ended duplex DNA is shown with heavy arrows. The type II cleavage sites yielding staggered cut is given by light arrows. For the type I restriction endonuclease recognition, the duplex cut is at 1 kilobase away and identical copies of DNA sequence interval is 4096 kilobase pairs as the sequence is 6 base longs with 4 possibility positioned
(b) Interpretation:
Type II cleavage sites producing blunt-ended duplex DNA should be identified using heavy arrows.
Concept Introduction:
The restriction site also known as the restriction sequence is an enzyme that splits DNA into fragments at a recognized site inside a molecule. The restriction site's other part is given and the type II cleavage site with the blunt-ended duplex DNA is shown with heavy arrows. The type II cleavage sites yielding staggered cut is given by light arrows. For the type I restriction endonuclease recognition, the duplex cut is at 1 kilobase away and identical copies of DNA sequence interval is 4096 kilobase pairs as the sequence is 6 base longs with 4 possibility positioned nucleotides.
(c) Interpretation:
Type II cleavage sites producing staggered cuts converted to recombinant DNA molecules by DNA ligase without enzyme involvement should be identified using light arrows.
Concept Introduction:
The restriction site also known as the restriction sequence is an enzyme that splits DNA into fragments at a recognized site inside a molecule. The restriction site's other part is given and the type II cleavage site with the blunt-ended duplex DNA is shown with heavy arrows. The type II cleavage sites yielding staggered cut is given by light arrows. For the type I restriction endonuclease recognition, the duplex cut is at 1 kilobase away and identical copies of DNA sequence interval is 4096 kilobase pairs as the sequence is 6 base longs with 4 possibility positioned nucleotides.
(d) Interpretation:
The place where the duplex be cut when the recognition site for type I restriction endonuclease should be identified.
Concept Introduction:
The restriction site also known as the restriction sequence is an enzyme that splits DNA into fragments at a recognized site inside a molecule. The restriction site's other part is given and the type II cleavage site with the blunt-ended duplex DNA is shown with heavy arrows. The type II cleavage sites yielding staggered cut is given by light arrows. For the type I restriction endonuclease recognition, the duplex cut is at 1 kilobase away and identical copies of DNA sequence interval is 4096 kilobase pairs as the sequence is 6 base longs with 4 possibility positioned nucleotides.
(e) Interpretation:
The average interval of identical copies of the given sequence in DNA which is random should be identified in kilobase pairs.
Want to see the full answer?
Check out a sample textbook solution
Chapter 21 Solutions
Biochemistry: Concepts and Connections
- what is a protein that contains a b-sheet and how does the secondary structure contributes to the overall function of the protein.arrow_forwarddraw and annotate a b-sheet and lable the hydrogen bonding. what is an example that contains the b-sheet and how the secondary structure contributes to the overall function of your example protein.arrow_forwardFour distinct classes of interactions (inter and intramolecular forces) contribute to a protein's tertiary and quaternary structures. Name the interaction then describe the amino acids that can form this type of interaction. Draw and annotate a diagram of the interaction between two amino acids.arrow_forward
- Examine the metabolic pathway. The enzymes that catalyze each step are identified as "e" with a numeric subscript. e₁ e3 e4 A B с 1° B' 02 e5 e6 e7 E F Which enzymes catalyze irreversible reactions? ப e ez ☐ ez e4 ☐ ப es 26 5 e7 Which of the enzymes is likely to be the allosteric enzyme that controls the synthesis of G? €2 ез e4 es 26 5 e7arrow_forwardAn allosteric enzyme that follows the concerted model has an allosteric coefficient (T/R) of 300 in the absence of substrate. Suppose that a mutation reversed the ratio. Select the effects this mutation will have on the relationship between the rate of the reaction (V) and substrate concentration, [S]. ㅁㅁㅁ The enzyme would likely follow Michaelis-Menten kinetics. The plot of V versus [S] would be sigmoidal. The enzyme would mostly be in the T form. The plot of V versus [S] would be hyperbolic. The enzyme would be more active.arrow_forwardPenicillin is hydrolyzed and thereby rendered inactive by penicillinase (also known as ẞ-lactamase), an enzyme present in some penicillin-resistant bacteria. The mass of this enzyme in Staphylococcus aureus is 29.6 kDa. The amount of penicillin hydrolyzed in 1 minute in a 10.0 mL. solution containing 1.00 x 10 g of purified penicillinase was measured as a function of the concentration of penicillin. Assume that the concentration of penicillin does not change appreciably during the assay. Plots of V versus [S] and 1/V versus 1/[S] for these data are shown. Vo (* 10 M minute"¹) 7.0 6.0 5.0 4.0 3.0 20 1.0 0.0 о 10 20 30 1/Vo (* 10 M1 minute) 20 103 90 BO 70 50 [S] (* 100 M) 40 50 60 y=762x+1.46 × 10" [Penicillin] (M) Amount hydrolyzed (uM) 1 0.11 3 0.25 5 0.34 10 0.45 30 0.58 50 0.61arrow_forward
- Consider the four graphs shown. In each graph, the solid blue curve represents the unmodified allosteric enzyme and the dashed green curve represents the enzyme in the presence of the effector. Identify which graphs correctly illustrate the effect of a negative modifier (allosteric inhibitor) and a positive modifier (allosteric activator) on the velocity curve of an allosteric enzyme. Place the correct graph in the set of axes for each type of modifier. Negative modifier Reaction velocity - Positive modifier Substrate concentration - Reaction velocity →→→→ Substrate concentration Answer Bankarrow_forwardConsider the reaction: phosphoglucoisomerase Glucose 6-phosphate: glucose 1-phosphate After reactant and product were mixed and allowed to reach at 25 °C, the concentration of each compound at equilibrium was measured: [Glucose 1-phosphate] = 0.01 M [Glucose 6-phosphate] = 0.19 M Calculate Keq and AG°'. Код .0526 Incorrect Answer 7.30 AG°' kJ mol-1 Incorrect Answerarrow_forwardClassify each phrase as describing kinases, phosphatases, neither, or both. Kinases Phosphatases Neither Both Answer Bank transfer phosphoryl groups to acidic amino acids in eukaryotes may use ATP as a phosphoryl group donor remove phosphoryl groups from proteins catalyze reactions that are the reverse of dephosphorylation reactions regulate the activity of other proteins catalyze phosphorylation reactions PKA as an example turn off signaling pathways triggered by kinasesarrow_forward
- Consider the reaction. kp S P kg What effects are produced by an enzyme on the general reaction? AG for the reaction increases. The rate constant for the reverse reaction (kr) increases. The reaction equilibrium is shifted toward the products. The concentration of the reactants is increased. The activation energy for the reaction is lowered. The formation of the transition state is promoted.arrow_forwardThe graph displays the activities of wild-type and several mutated forms of subtilisin on a logarithmic scale. The mutations are identified as: • The first letter is the one-letter abbreviation for the amino acid being altered. • The number identifies the position of the residue in the primary structure. ⚫ The second letter is the one-letter abbreviation for the amino acid replacing the original one. • Uncat. refers to the estimated rate for the uncatalyzed reaction. Log₁(S-1) Wild type S221A H64A -5 D32A S221A H64A D32A -10 Uncat. How would the activity of a reaction catalyzed by a version of subtilisin with all three residues in the catalytic triad mutated compare to the activity of the uncatalyzed reaction? It would have more activity, because the reaction catalyzed by the triple mutant is approximately three-fold faster than the uncatalyzed reaction. It would have less activity, because the reaction catalyzed by the triple mutant is approximately 1000-fold slower than the…arrow_forwardB Substrate Product AL Product Substrate Reaction progress- Reaction progress- omplete the passage describing the two reactions. In reaction A, the stability of the substrate is (AG) of the reaction is positive, Incorrect Answer greater than the stability of the product. The free-energy change Incorrect Answer so the reaction is considered In reaction B, the stability of the substrate is (AG) of the reaction is less than Incorrect Answer endergonic and Incorrect Answer not spontaneous. Incorrect Answer the stability of the product. The free-energy change negative, so the reaction is considered Incorrect Answer exergonic and spontaneous. Incorrect Answer Incorrect Answerarrow_forward
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON





