
Concept explainers
(a)
To draw:
The diagram that consist of 5 genes and the enhancers. Suppose that the blue, orange, green, red, purple and black are the activator proteins that binds to the appropriate color-coded control elements of the enhancers.
(a)
To draw an X above the enhancer elements that will transcribe only gene 5 and also, identify the colored activators present.
(b)
To draw a dot above the enhancer elements where blue, orange and green activators are present. Also, identify the transcribed genes.
(c)
Suppose, the genes 1, 2 and 4 codes for the proteins that are nerve-specific and genes 3 and 5 are the proteins that are skin-specific. To identify the activators that will ensure transcription of the appropriate genes.
Concept introduction:
A gene (eukaryotic) and the elements of DNA (deoxyribonucleic acid) are organized properly. The transcription initiation complex (cluster of proteins) assembles on the promoter sequence and the processing or transcription process is initiated.
Pictorial representation:
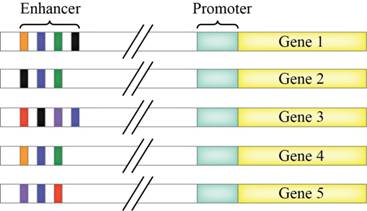
Figure 1: Transcription of genes with the help of enhancer elements.
To draw:
An X above the enhancer elements that will transcribe only gene 5 and also, identify the colored activators present.
(b)
To draw:
A dot above the enhancer elements where blue, orange and green activators are present. Also, identify the associated transcribed genes.
(c)
To suppose:
The genes 1, 2 and 4 codes for the proteins that are nerve-specific and genes 3 and 5 codes for the proteins that are skin-specific. And, identify the activators that will ensure transcription of the appropriate genes.
Want to see the full answer?
Check out a sample textbook solution
Chapter 15 Solutions
Campbell Biology in Focus, Books a la Carte Edition; Modified Mastering Biology with Pearson eText - ValuePack Access Card - for Campbell Biology in Focus (2nd Edition)
- answer questions 1-10arrow_forwardAnswer Question 1-9arrow_forwardEx: Mr. Mandarich wanted to see if the color of light shined on a planthad an effect on the number of leaves it had. He gathered a group ofthe same species of plants, gave them the same amount of water, anddid the test for the same amount of time. Only the color of light waschanged. IV:DV:Constants:Control Gr:arrow_forward
- ethical considerations in medical imagingarrow_forwardPlease correct answer and don't used hand raiting and don't used Ai solutionarrow_forward2. In one of the reactions of the citric acid cycle, malate is oxidized to oxaloacetate. When this reaction is considered in isolation, a small amount of malate remains and is not oxidized. The best term to explain this is a. enthalpy b. entropy c. equilibrium d. free energy e. loss of energyarrow_forward
- 18. The citric acid cycle takes place in a. the chloroplasts b. the cytosol c. the inner mitochondrial membrane d. between the two mitochondrial membranes e. the mitochondrial matrix 40 WILarrow_forward8. Most reactions of anaerobic respiration are similar to a. aerobic respiration b. photosynthesis c. lactic acid fermentation d. alcoholic fermentation e. both c and darrow_forward12. Which of the following molecules can absorb light? a. Pigments b. Chlorophyll c. Rhodopsin d. Carotenoids e. All of the abovearrow_forward
- Which of the following proteins or protein complexes is directly required for the targeting of mitochondrial inner membrane multipass proteins, such as metabolite transporters, whose signal sequence is normally not cleaved after import? OA. TIM22 OB. TIM23 C. OXA OD. Mia40 OE SAMarrow_forwardQUESTION 9 An animal cell has been wounded and has a small rupture in its plasma membrane. Which of the following is more likely to happen next? OA. The cell rapidly cleaves by cytokinesis. OB. The rate of receptor-mediati endocytosis is increased. OC. The rate of exocytosis is increased. OD. The rate of pinocytosis is increased.arrow_forwardFor the a subunit of a trimeric G protein, A. a G-protein-coupled receptor GPCR) acts as a guanine nucleotide exchange factor (GEF), whereas a regulator of G protein signaling (RGS) can act as a GTPase-activating protein (GAP). B. a GPCR acts as a GAP, whereas an RGS can act as a GEF. C. both a GPCR and an RGS can act as a GEF. O D. both a GPCR and an RGS can act as a GAP OE. None of the above.arrow_forward
 Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
 Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning





