
Concept explainers
To estimate: The K value from the given data that describes the ligand binding to a protein.
Concept introduction: Dissociation constant is the equilibrium constant at which the ligand dissociates from the protein. It is denoted as K. Y is fractional saturation of the ligand occupying the ligand-binding site in the protein, or it can be defined as the number of ligands that has dissociated from the protein. K is achieved when fractional saturation is half at a particular ligand concentration.
Answer to Problem 1E
Answer: The dissociation constant is estimated by plotting the values in a graph and finding the ligand concentration when Y is 0.5 (half of fractional saturation).
Pictorial representation:
Fig.1 shows a graph with ligand (mM) in the X-axis and Y in the Y-axis.
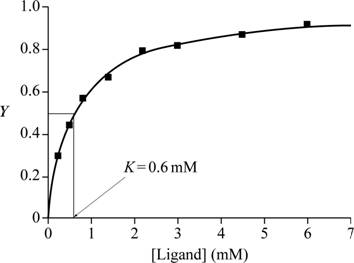
Fig.1: Graph of ligand (mM) vs Y to estimate K.
Explanation of Solution
Explanation:
When fractional saturation is half, it means that the ligand has occupied half of the ligand binding site in the protein. When fractional saturation is half, half of the ligand is bound to half the number of proteins in the medium. Half of proteins and half of ligands are free in the medium. This can be called as the equilibrium state of the ligand-protein binding reaction, and thus K is obtained at half of the fractional saturation.
A graph is plotted with ligand concentration in mM on the X-axis and Y, the fractional saturation, on the Y-axis with the given data. At half of fractional saturation, that is, at equilibrium state, the corresponding ligand concentration is 0.6 Mm, which is evident from the graph.
Conclusion: From the graph plotted for the given values, the K value is 0.6mM.
Want to see more full solutions like this?
Chapter 7 Solutions
FUNDAMENTALS OF BIOCHEMISTRY-ACCESS
- Draw the reaction between sphingosine and arachidonic acid. Draw out the full structures.arrow_forwardDraw both cis and trans oleic acid. Explain why cis-oleic acid has a melting point of 13.4°C and trans-oleic acid has a melting point of 44.5°C.arrow_forwardDraw the full structure of the mixed triacylglycerol formed by the reaction of glycerol and the fatty acids arachidic, lauric and trans-palmitoleic. Draw the line structure.arrow_forward
- Draw out the structure for lycopene and label each isoprene unit. "Where is lycopene found in nature and what health benefits does it provide?arrow_forwardWhat does it mean to be an essential fatty acid? What are the essential fatty acids?arrow_forwardCompare and contrast primary and secondary active transport mechanisms in terms of energy utilisation and efficiency. Provide examples of each and discuss their physiological significance in maintaining ionic balance and nutrient uptake. Rubric Understanding the key concepts (clearly and accurately explains primary and secondary active transport mechanisms, showing a deep understanding of their roles) Energy utilisation analysis ( thoroughly compares energy utilisation in primary and secondary transport with specific and relevant examples Efficiency discussion Use of examples (provides relevant and accurate examples (e.g sodium potassium pump, SGLT1) with clear links to physiological significance. Clarity and structure (presents ideas logically and cohesively with clear organisation and smooth transition between sections)arrow_forward
- 9. Which one of the compounds below is the major organic product obtained from the following reaction sequence, starting with ethyl acetoacetate? 요요. 1. NaOCH2CH3 CH3CH2OH 1. NaOH, H₂O 2. H3O+ 3. A OCH2CH3 2. ethyl acetoacetate ii A 3. H3O+ OH B C D Earrow_forward7. Only one of the following ketones cannot be made via an acetoacetic ester synthesis. Which one is it? Ph کہ A B C D Earrow_forward2. Which one is the major organic product obtained from the following reaction sequence? HO A OH 1. NaOEt, EtOH 1. LiAlH4 EtO OEt 2. H3O+ 2. H3O+ OH B OH OH C -OH HO -OH OH D E .CO₂Etarrow_forward
- what is a protein that contains a b-sheet and how does the secondary structure contributes to the overall function of the protein.arrow_forwarddraw and annotate a b-sheet and lable the hydrogen bonding. what is an example that contains the b-sheet and how the secondary structure contributes to the overall function of your example protein.arrow_forwardFour distinct classes of interactions (inter and intramolecular forces) contribute to a protein's tertiary and quaternary structures. Name the interaction then describe the amino acids that can form this type of interaction. Draw and annotate a diagram of the interaction between two amino acids.arrow_forward
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON





