
Figure 28.3 Which of the following statements is
false?
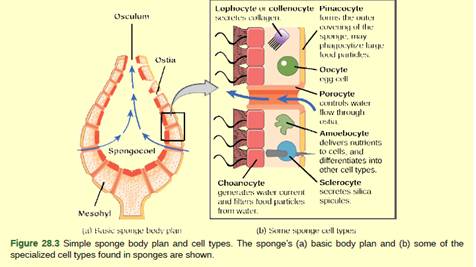
- Choanocytes have flagella that propel water through the body.
Introduction:
Generally, sponges do not show true tissue-layer organization and do not have a number of functional tissues, that made up of different cell types specialized for distinct functions such as pinacocytes Pinacocytes is epithelial-like cells and form the outermost body, called a pinacoderm, that serves a protective function similar to the epidermis. Pinacoderm scattered among all area of Ostia that allow entry of water into the body of the sponge.
Answer to Problem 1VCQ
Correct answer:
Correct answer is option (b), Pinacocytes can transform into any cell type.
Explanation of Solution
Explanation/justification for the correct answer:
Option (b) is given as Pinacocytes can transform into any cell type. Pinacoderm scattered among all area of ostia that allow entry of water into the body of the sponge. In some sponges, ostia are made up of porocytes, these single tube-shaped cells act as valves to regulate the flow of water into the spongocoel. It is a correct answer.
Explanation for the incorrect answer:
Option (a) is given as choanocytes have flagella that propel water through the body. Choanocytes line is the inner portions of spaces in the sponges that allow the flow of water. The water current is maintained through the presence of flagella so that food particles can be trapped and ingested via phagocytosis. It is a wrong answer.
Option (c) is given as lymphocytes secret collagen. Lophocytes is a specific type of cells in sponges that able to produce protein like collagen for maintenance of mesophyll. it is a wrong answer.
Option (d) is given as porocytes control the flow of the water through pores in the sponge. Porifera is a phylum that cover sponges that pore-bearers, in some sponges, porocytes make ostia like single tube-shaped cells that act as valves and help to regulate the flow of water into the spongocoel. it is a wrong answer.
Thus, Pinacocytes can transform into any cell type is a false statement.
Want to see more full solutions like this?
Chapter 28 Solutions
BIOLOGY FOR AP COURSES (OER)
Additional Science Textbook Solutions
Physics for Scientists and Engineers: A Strategic Approach, Vol. 1 (Chs 1-21) (4th Edition)
Introductory Chemistry (6th Edition)
Genetic Analysis: An Integrated Approach (3rd Edition)
Biology: Life on Earth (11th Edition)
Microbiology: An Introduction
Biological Science (6th Edition)
- Please indentify the unknown organismarrow_forwardPlease indentify the unknown organismarrow_forward5G JA ATTC 3 3 CTIA A1G5 5 GAAT I I3 3 CTIA AA5 Fig. 5-3: The Eco restriction site (left) would be cleaved at the locations indicated by the arrows. However, a SNP in the position shown in gray (right) would prevent cleavage at this site by EcoRI One of the SNPs in B. rapa is found within the Park14 locus and can be detected by RFLP analysis. The CT polymorphism is found in the intron of the Bra013780 gene found on Chromosome 1. The Park14 allele with the "C" in the SNP has two EcoRI sites and thus is cleaved twice by EcoRI If there is a "T" in that SNP, one of the EcoRI sites is altered, so the Park14 allele with the T in the SNP has only one EcoRI site (Fig. 5-3). Park14 allele with SNP(C) Park14 allele with SNPT) 839 EcoRI 1101 EcoRI 839 EcoRI Fig. 5.4: Schematic restriction maps of the two different Park14 alleles (1316 bp long) of B. rapa. Where on these maps is the CT SNP located? 90 The primers used to amplify the DNA at the Park14 locus (see Fig. 5 and Table 3 of Slankster et…arrow_forward
- From your previous experiment, you found that this enhancer activates stripe 2 of eve expression. When you sequence this enhancer you find several binding sites for the gap gene, Giant. To test how Giant interacts with eve, you decide to remove all of the Giant binding sites from the eve enhancer. What results do you expect to see with respect to eve expression?arrow_forwardWhat experiment could you do to see if the maternal gene, bicoid, is sufficient to form anterior fates?arrow_forwardYou’re curious about the effect that gap genes have on the pair-rule gene, evenskipped (eve), so you isolate and sequence each of the eve enhancers. You’re particularly interested in one of the enhancers, which is just upstream of the eve gene. Describe an experimental technique you would use to find out where this particular eve enhancer is active.arrow_forward
- For short answer questions, write your answers on the line provided. To the right is the mRNA codon table to use as needed throughout the exam. First letter U บบบ U CA UUCPhe UUA UCU Phe UCC UUG Leu CUU UAU. G U UAC TV UGCys UAA Stop UGA Stop A UAG Stop UGG Trp Ser UCA UCG CCU] 0 CUC CUA CCC CAC CAU His CGU CGC Leu Pro CCA CAA Gin CGA Arg CUG CCG CAG CGG AUU ACU AAU T AUC lle A 1 ACC Thr AUA ACA AUG Mot ACG AGG Arg GUU GCU GUC GCC G Val Ala GAC Asp GGU GGC GUA GUG GCA GCG GAA GGA Gly Glu GAGJ GGG AACASH AGU Ser AAA1 AAG Lys GAU AGA CAL CALUCAO CAO G Third letter 1. (+7) Use the table below to answer the questions; use the codon table above to assist you. The promoter sequence of DNA is on the LEFT. You do not need to fill in the entire table. Assume we are in the middle of a gene sequence (no need to find a start codon). DNA 1 DNA 2 mRNA tRNA Polypeptide C Val G C. T A C a. On which strand of DNA is the template strand (DNA 1 or 2)?_ b. On which side of the mRNA is the 5' end (left or…arrow_forward3. (6 pts) Fill in the boxes according to the directions on the right. Structure R-C R-COOH OH R-OH i R-CO-R' R R-PO4 R-CH3 C. 0 R' R-O-P-OH 1 OH H R-C-H R-N' I- H H R-NH₂ \H Name Propertiesarrow_forward4. (6 pts) Use the molecule below to answer these questions and identify the side chains and ends. Please use tidy boxes to indicate parts and write the letter labels within that box. a. How many monomer subunits are shown? b. Box a Polar but non-ionizable side chain and label P c. Box a Basic Polar side chain and label BP d. Box the carboxyl group at the end of the polypeptide and label with letter C (C-terminus) H H OHHO H H 0 HHO H-N-CC-N-C-C N-C-C-N-GC-OH I H-C-H CH2 CH2 CH2 H3C-C+H CH2 CH2 OH CH CH₂ C=O OH CH2 NH2arrow_forward
 Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Human Physiology: From Cells to Systems (MindTap ...BiologyISBN:9781285866932Author:Lauralee SherwoodPublisher:Cengage Learning
Human Physiology: From Cells to Systems (MindTap ...BiologyISBN:9781285866932Author:Lauralee SherwoodPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning





