
Concept explainers
(a)
To determine: The range of [IPTG] expression level which vary from 10% to 90%
Introduction:
Isopropyl β-D-1-thiogalactopyranoside (IPTG) binds to the lac repressor in similar way as allolactose. It releases the repressor in allosteric manner from the lac operator and the structural gene transcription of lac operon.
(a)
Explanation of Solution
Explanation:
10% expression of [IPTG] also represents 90% repression of. [IPTG] Hence, 10% of the repressor will show binding to the inducer, while remaining 90% of the repressor will be available for binding to the operator.
To calculate the range of 10% expression level of, the [IPTG] following formula is used with reference to equation 5-8.
So the range of expression level of [IPTG] is:
To calculate the range of 90% expression level of [IPTG],
So the range of expression level of [IPTG] is:
The range of expression of proteins of lac operon varies from 10-fold over the 10-fold expression of [IPTG] range.
The range of IPTG expression level varies from 10-90% or varies 10 folds.
(b)
To describe: The level of lac operon proteins present in E. coli cell before, during, and after IPTG induction.
Introduction:
Isopropyl β-D-1-thiogalactopyranoside (IPTG) binds to the lac repressor in similar way as allolactose. It releases the repressor in allosteric manner from the lac operator and the structural gene transcription of lac operon.
(b)
Explanation of Solution
Explanation:
IPTG induces the expression of lac operon proteins. The level of lac protein is very low before the induction, and the expression of lac protein will increase during the induction. After induction, the proteins synthesis will stop thereby, decreasing the expression of lac proteins. The lac proteins will increase during induction, and it decreases before and after the induction.
(c)
To explain: The lac operon lacks both the characteristics: A. that is either fully on or fully off, B. both states are stable.
Introduction:
In normal lac operon regulation, when the repressor is bound to lactose (inducer), it no longer binds to the operator DNA. Binding of lactose to the repressor alters the conformation of repressor protein so that it does not have affinity for the operator. The change in the shape of repressor gene will not allow it to bind to lactose resulting in inhibition of translation of structural gene.
(c)
Explanation of Solution
Explanation:
Lac operon has multiple expression levels, other than the on or off condition, as condition A is defined by only two states that are fully on or fully off, it is not followed by lac operon.
Lac operon expression depends on the binding of inducer molecule, as the removal of inducer decreases the expression of lac operon. Condition B is defined by “both states are stable” is not followed by lac operon.
Lac operon shows different expressions thus it cannot be fully on or fully off. It is also not present in stable condition as its expression depends on inducer molecule.
(d)
To explain: The proteins that are present and which promoter is being expressed for the states GFP-on and GFP-off
Introduction:
In normal lac operon regulation, when the repressor is bound to lactose (inducer), it no longer binds to the operator DNA. Binding of lactose to the repressor alters the conformation of the repressor protein so that it does not have affinity for the operator. The change in shape of repressor gene will not allow it to bind to lactose resulting in inhibition of translation of structural gene.
(d)
Explanation of Solution
Pictorial representation: Fig.1 represents the Plasmid (for the states GFP-on and GFP-ff)
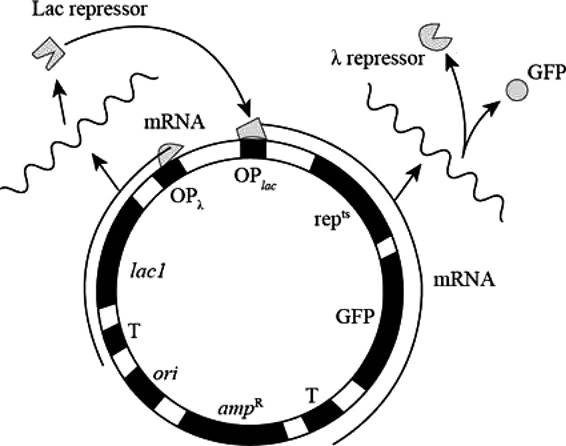
Fig. 1: Plasmid (for the states GFP-on and GFP-off )
Explanation:
GFP-on: The repts gene expresses the repts protein in high level which represses the expression of OPλ, and GFP are expressed at high level during GFP-on state.
GFP-off: high level expression of Lac I represses the expression of OPlac, repts and GFP.
In GFP-on state repts and GFP proteins are present, and in GFP-off state Lac I protein is present.
(e)
To determine: The state from which IPTG would toggle the system
Introduction:
IPTG induces the expression of lac operon proteins. The level of lac protein is very low before the induction while the expression of lac protein will increase during the induction. After the induction, the proteins synthesis will stop thereby decreasing the expression of lac proteins.
(e)
Explanation of Solution
Explanation:
Presence of LacI protein enables the system for induction of IPTG, thus it induces the expression only in GFP-off state, and the induction is followed by expression of OPlac, repts and GFP. Thus, IPTG toggle the system from GFP-off to GFP-on.
The state from which IPTG would toggle the system is from GFP-off to GFP-on.
(f)
To determine: The state from which treatment with heat (42°C) would toggle the system.
Introduction:
In normal lac operon regulation, when the repressor is bound to lactose (inducer), it no longer binds to the operator DNA. Binding of lactose to the repressor alters the conformation of the repressor protein so that it does not have affinity for the operator. The change in shape of repressor gene will not allow it to bind to lactose resulting in inhibition of translation of structural gene.
(f)
Explanation of Solution
Explanation:
Heat affects the system only in presence of repts, which is expressed only in GFP-on state. The repts which is inactivated due to heat exposure results in OPλ repression followed by the expression of lac I repressor which represses the GFP expression. Hence, heat toggles the system from GFP-on to GFP-off.
(g)
To determine: The reason that plasmid has characteristics A and B.
Introduction:
In normal lac operon regulation, when the repressor is bound to lactose (inducer), it no longer binds to the operator DNA. Binding of lactose to the repressor alters the conformation of the repressor protein so that it does not have affinity for the operator. The change in shape of repressor gene will not allow it to bind to lactose resulting in inhibition of translation of structural gene.
(g)
Explanation of Solution
Explanation:
Plasmid system has two states of expression. In intermediate state, the system is not stable hence the plasmid follows the condition A which defines the only on and off condition.
Plasmid remains in a single state even after the removal of repressor from binding site, and as condition B defines that both the states are stable, it is followed by the plasmid.
Plasmid has characteristics A and B as it has only two states of expression, and it is stable in both states.
(h)
To explain: The characteristic of system A that demonstrates GFP expression level in individual cells at
Introduction:
IPTG induces the expression of lac operon proteins. The level of lac protein is very low before the induction while the expression of lac protein will increase during the induction. After the induction, the protein synthesis will stop thereby decreasing the expression of lac proteins.
(h)
Explanation of Solution
Explanation:
Characters of system A is defined by only on or only off state, at intermediate concentration of inducer X. In the system, some cells are GFP-on and some are GFP-off, hence they define the system A.
As the cell population have both type GFP-on and GFP-off cells in the system, the expression level of
The cells’ population contains only GFP-on and GFP-off cells in the system. It defines the condition A. As two types of expression are there in the system, it shows bimodal distribution of expression level of
Want to see more full solutions like this?
Chapter 28 Solutions
SaplingPlus for Lehninger Principles of Biochemistry (Six-Month Access)
- Please draw out the mechanism with curved arrows showing electron flow. Pyruvate is accepted into the TCA cycle by a “feeder” reaction using the pyruvate dehydrogenase complex, resulting in acetyl-CoA and CO2. Provide the mechanism for this reaction utilizing the TPP cofactor. Include the roles of all cofactors.arrow_forwardPyruvate is accepted into the TCA cycle by a “feeder” reaction using the pyruvate dehydrogenase complex, resulting in acetyl-CoA and CO2. Provide the mechanism for this reaction utilizing the TPP cofactor. Include the roles of all cofactors.arrow_forwardThe mitochondrial ATP synthase has 10 copies of the F0 subunit “c”, and the [H ] in the mitochondrial inner membrane space (IMS) is 6.31 x 10-8 M and the [H + ] in the matrix is 3.16 x 10-9 M. Calculate the minimum membrane potential (∆Ψ) necessary to make ATP synthesis thermodynamically favorable. [Assume ∆G' ofphosphate hydrolysis of ATP is - 45 kJ/mol.]arrow_forward
- B- Vitamins are converted readily into important metabolic cofactors. Deficiency in any one of them has serious side effects. a. The disease beriberi results from a vitamin B 1 (Thiamine) deficiency and is characterized by cardiac and neurological symptoms. One key diagnostic for this disease is an increased level of pyruvate and α-ketoglutarate in the bloodstream. How does this vitamin deficiency lead to increased serumlevels of these factors? b. What would you expect the effect on the TCA intermediates for a patient suffering from vitamin B 5 deficiency? c. What would you expect the effect on the TCA intermediates for a patientsuffering from vitamin B 2 /B 3 deficiency?arrow_forwardPyruvate is accepted into the TCA cycle by a “feeder” reaction using the pyruvate dehydrogenase complex, resulting in acetyl-CoA and CO2. Provide a full mechanism for this reaction utilizing the TPP cofactor. Include the roles of all cofactors.arrow_forwardMap out all of the metabolic pathways in the liver cell. Draw out the structures and names of all compounds neatly by hand and the pathways responsible for metabolizing them. Some examples are: Glycolysis/gluconeogenesis, PPP, Glycogenesis/glycogenolysis, Krebs, ETC, selectamino acid pathways (Ala, Glu, Asp) Lipogenesis/lipolysis. Citrate/MAS/glycerol phosphate shuttlesystems, and the Cori/Glc-Ala cycles. Rules:-Draw both a mitochondrial area of metabolism and a cytoplasmic area of metabolism.-Draw the liver and its roles in glucose recycling (Cori cycle/Glc-Alanine recycling)-Avoid drawing the same molecule twice (except for separate mitochondrial/cytoplasmic populations. i.e. Design the PPP/Glycolysis so that GAP is only drawn once)-Label Carbon 4 of glucose and highlight where you would expect to find it in EVERY compound in whichit is present.-Have one or two locations for NADH/NADPH/ATP/GTP/CoQH2 – many arrows will come to/from thesespots.arrow_forward
- a. Draw the Krebs Cycle and show the entry points for the amino acids Alanine,Glutamic Acid, Asparagine, and Valine into the Krebs Cycle. (Include name of Enzymes involved) b. How many rounds of Krebs will be required to waste all Carbons of Glutamic Acid as CO2? (Show by drawing out the mechanism that occurs)arrow_forwardThe malate-aspartate shuttle allows malate to be exchanged for aspartate acrossthe inner mitochondrial membrane. (a) Describe the role of the malate-aspartate shuttle in liver cells under HIGHblood glucose conditions. Be sure to explain your answer. (b) Describe the role of the malate-aspartate shuttle in liver cells under LOW blood + glucose conditions.arrow_forward(a) Write out the net reaction, calculate ∆E ̊' for the reaction, and calculate the standard free-energy change (∆G°') for the overall oxidation/reduction reaction. (h) How many moles of ATP could theoretically be generated per mole of FADH2 oxidized by this reaction, given a ∆G ̊' of ATP synthesis of + 31 kJ/mol? How many moles of ATP could be generated per mole of FADH2 oxidized by this reaction under more typical cellular conditions (where ∆G' of ATP hydrolysis is ~ -50 kJ/mol)? Be sure to show your work and explain your answer.arrow_forward
- Indicate for the reactions below which type of enzyme and cofactor(s) (if any) would be required to catalyze each reaction shown. 1) Fru-6-P + Ery-4-P <--> GAP + Sed-7-P2) Fru-6-P + Pi <--> Fru-1,6-BP + H2O3) GTP + ADP <--> GDP + ATP4) Sed-7-P + GAP <--> Rib-5-P + Xyl-5-P5) Oxaloacetate + GTP ---> PEP + GDP + CO26) DHAP + Ery-4-P <--> Sed-1,7-BP + H2O7) Pyruvate + ATP + HCO3- ---> Oxaloacetate + ADP + Piarrow_forwardThe phosphate translocase is an inner mitochondrial membrane symporter that transports H2PO4- and H+ into the mitochondrial matrix. Phosphate is a substrate for Complex V (the ATP Synthase), the enzyme that couples the synthesis of ATP to the H+ gradient formed by the electron transport chain. (a) Bongotoxin is a hypothetical compound that inhibits the phosphate translocase of the inner mitochondrial membrane. Explain why electron transport from NADH to O2 stops when bongotoxin is added to mitochondria (i.e., why do electrons stop flowing through the electron transport chain even with an abundance of NADH and O 2 present). What effect will the addition of the weak acid dinitrophenol (DNP) to the cytosol have on electron transport in bongotoxin-inhibited mitochondria? Be sure to explain your answers. (b) How much free energy is released (in kJ) when one mole of protons flows from the mitochondrial inner membrane space (IMS) to the mitochondrial matrix when the [H+ ] in the IMS is 7.9 x…arrow_forwardWhen TMPD/ascorbate is added to mitochondria as a source of electrons (TMPD/ascorbate reduce cytochrome c directly) oxygen is reduced to H2O by the electron transport chain (ETC).(a) Approximately how many ATPs would result per O2 consumed when electrons come from TMPD/ascorbate? (b) If dinitrophenol (DNP) is added to the mitochondria in (a) above, what effect would DNP have on the yield of ATPs per O2 reduced from TMPD/ascorbate electrons?arrow_forward
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON





