
Concept explainers
(a)
To explain: The way through which nondisjunction of the synthetic chromosome would give rise to a colony that is half red and half white.
Introduction:
Yeast artificial chromosomes (YACs) clone large pieces of deoxyribonucleic acid (DNA) in yeast cells. It can generate unlimited number of DNA copies. Cloning genomic DNA into YAC is useful to analyze very long DNA sequence such as human genes.
The three types of DNA sequence that are necessary to ensure proper propagation and replication of a YAC in a yeast cell are as follows:
1) Centromere for the attachment of mitotic spindle fibers. It attracts one copy of each replicated chromosome into each new daughter cell.
2) Telomere is found at each end of a chromosome. It prevents the linear DNA from degradation.
3) Autonomous replicating sequence (origin of replication) allows assembly of
(a)
Explanation of Solution
In the given experiment, Heiter and colleagues constructed synthetic chromosomes to find out the components of yeast chromosomes. They constructed plasmids with different parts of chromosomes. They observed proper segregation of synthetic chromosomes during mitosis. According to the assay, wild-type yeast colonies are white and mutated copies of adenine-requiring yeast colonies are red.
During non-disjunction of synthetic chromosomes, one daughter cell and its entire descendants obtain two copies of the synthetic chromosome and give rise to colonies that are white. On the other hand, other daughter cell and all of its descendants obtain no copies of the synthetic chromosome and give rise to colonies that are red. This leads the formation of half-white, half-red colony.
(b)
To explain: The way through which loss of the synthetic chromosome would give rise to a colony that is half red and half pink.
Introduction:
Yeast artificial chromosomes (YACs) clone large pieces of DNA in yeast cells. It can generate unlimited number of DNA copies. Cloning genomic DNA into YAC is useful to analyze very long DNA sequence such as human genes.
The three types of DNA sequence that are necessary to ensure proper propagation and replication of a YAC in a yeast cell are as follows:
1) Centromere for the attachment of mitotic spindle fibers. It attracts one copy of each replicated chromosome into each new daughter cell.
2) Telomere is found at each end of a chromosome. It prevents the linear DNA from degradation.
3) Autonomous replicating sequence (origin of replication) allows assembly of DNA replication machinery and causes the movement of replication fork.
(b)
Explanation of Solution
If synthetic chromosome loss, one daughter cell and all of its descendants obtain one copy of the synthetic chromosome that give rise to form pink colony. The other daughter cell and all of its descendants obtain no copies of the synthetic chromosome and give rise to form red colony. Thus, a colony formed that is half-pink and half-red.
(c)
To determine: The size of the centromere necessary for normal mitotic segregation along with the reason.
Introduction:
Yeast artificial chromosomes (YACs) are necessary to clone large pieces of DNA in yeast cells. It can generate unlimited number of DNA copies. Cloning genomic DNA into YAC is useful to analyze very long DNA sequence such as human genes.
The three types of DNA sequence that are necessary to ensure proper propagation and replication of a YAC in a yeast cell are as follows:
1) Centromere for the attachment of mitotic spindle fibers. It attracts one copy of each replicated chromosome into each new daughter cell.
2) Telomere is found at each end of a chromosome. It prevents the linear DNA from degradation.
3) Autonomous replicating sequence (origin of replication) allows assembly of DNA replication machinery and causes the movement of replication fork.
(c)
Explanation of Solution
Heiter and colleagues estimated that the frequency of aberrant mitotic procedures with different types of synthetic chromosomes by counting the frequency of different colony types. They constructed synthetic chromosomes with DNA fragments of various sizes having a known centromere.
Based on the given data, it can be concluded that the centromere size needed for normal mitotic segregation should be less than 0.63 kbp. This is because all the fragments of DNA of 0.63 kbp size contain mitotic stability and cannot show loss of chromosomes.
Thus, the minimum functional centromere should be less than 0.63 kbp, because all fragments of this size or larger determine relative mitotic stability.
Thus, the centromere size required for normal mitotic segregation should be smaller than 0.63 kbp.
(d)
To explain: The way through which the synthetic chromosomes can be replicated more-or-less properly.
Introduction:
Yeast artificial chromosomes (YACs) are required to clone large pieces of DNA in yeast cells. It can generate unlimited number of DNA copies. Cloning genomic DNA into YAC is useful to analyze very long DNA sequence such as human genes.
The three types of DNA sequence that are necessary to ensure proper propagation and replication of a YAC in a yeast cell are as follows:
1) Centromere for the attachment of mitotic spindle fibers. It attracts one copy of each replicated chromosome into each new daughter cell.
2) Telomere is found at each end of a chromosome. It prevents the linear DNA from degradation.
3) Autonomous replicating sequence (origin of replication) allows assembly of DNA replication machinery and causes the movement of replication fork.
(d)
Explanation of Solution
Telomeres are ribonucleic proteins that are the “protective ends” of “linear chromosomes”. Telomeres generally shorten throughout the lifetime of a person. The synthetic chromosomes are constructed to determine the components of yeast chromosomes.
Telomeres are not required to replicate circular chromosomes beacuse telomeres are important to completely replicate linear DNA. Since, synthetic chromosomes are circular; hence, telomeres are not required for their replication
(e)
To explain: The chromosome size required for normal mitotic segregation and the reasons for it.
Introduction:
Yeast artificial chromosomes (YACs) are required to clone large pieces of DNA in yeast cells. It can generate unlimited number of DNA copies. Cloning genomic DNA into YAC is useful to analyze very long DNA sequence such as human genes.
The three types of DNA sequence that are necessary to ensure proper propagation and replication of a YAC in a yeast cell are as follows:
1) Centromere for the attachment of mitotic spindle fibers. It attracts one copy of each replicated chromosome into each new daughter cell.
2) Telomere is found at each end of a chromosome. It prevents the linear DNA from degradation.
3) Autonomous replicating sequence (origin of replication) allows assembly of DNA replication machinery and causes the movement of replication fork.
(e)
Explanation of Solution
According to given data, if chromosome size is larger, then there will be less chances of total error rate. Larger is the chromosome, the more closely it is segregated. According to the given data, neither a lesser size below, neither which the synthetic chromosome is fully unstable, nor does a greater size above, which stability wills no longer change.
(f)
To determine: Whether the centeromeric and telomeric sequences used in this experiment explain the mitotic stability of normal yeast chromosomes or must other elements be involved.
Introduction:
Yeast artificial chromosomes (YACs) are required to clone large pieces of DNA in yeast cells. It can generate unlimited number of DNA copies. Cloning genomic DNA into YAC is useful to analyze very long DNA sequence such as human genes.
The three types of DNA sequence that are necessary to ensure proper propagation and replication of a YAC in a yeast cell are as follows:
1) Centromere for the attachment of mitotic spindle fibers. It attracts one copy of each replicated chromosome into each new daughter cell.
2) Telomere is found at each end of a chromosome. It prevents the linear DNA from degradation.
3) Autonomous replicating sequence (origin of replication) allows assembly of DNA replication machinery and causes the movement of replication fork.
(f)
Explanation of Solution
Graphical representation:
Fig.1 represents the graphical representation of the length of DNA and total percentage of the error rate.
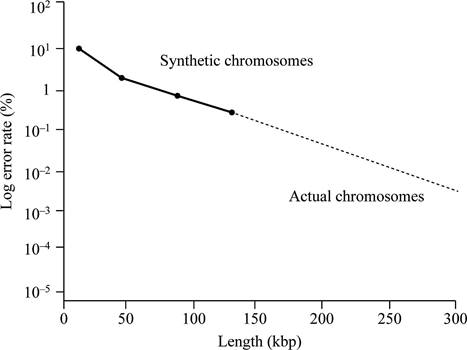
Fig.1: The length of DNA and total percentage of the error rate.
Normal yeast chromosomes are linear in structure and ranges from 250 kbp to 2,000 kbp. They have a mitotic error frequency of 10-5 per cell division. Based on the given data, a graph can be plotted. This graph is plotted against the length of DNA and total percentage of the error rate.
Hence, it can be concluded that the replication will not be stable even if the synthetic chromosomes are longer in size like normal yeast chromosomes. It is due to the requirement of some additional elements (other, as yet undiscovered, elements) are required for stability that occur in normal chromosomes.
Want to see more full solutions like this?
Chapter 24 Solutions
SAPLINGPLUS FOR PRINCIPLES OF BIOCHEMIS
- The reduced coenzymes generated by the citric acid cycle donate electrons in a series of reactions called the electron-transport chain. The energy from the electron-transport chain is used for oxidative phosphorylation. Which compounds donate electrons to the electron- transport chain? H₂O NADH பப NAD+ ATP ADP FADH₂ FAD Which compounds are the final products of the electron-transport chain and oxidative phosphorylation? H₂O NADH NAD+ ΠΑΤΡ Π ADP FADH₂ FAD Which compound is the final electron acceptor in the electron-transport chain? Оно NADH NAD+ ATP ADP FADH₂ FADarrow_forwardHexokinase in red blood cells has a Michaelis constant (KM) of approximately 50 μM. Because life is hard enough as it is, let's assume that hexokinase displays Michaelis-Menten kinetics. What concentration of blood glucose yields an initial velocity (V) equal to 90% of the maximal velocity (Vmax)? [glucose] = What does the calculated substrate concentration at 90% Vmax tell you if normal blood glucose levels range between approximately 3.6 and 6.1 mM? Hexokinase operates near Vmax only when glucose levels are low. Hexokinase normally operates far below Vmax. Hexokinase operates near Vmax only when glucose levels are high. Hexokinase normally operates near Vmax mMarrow_forwardClassify each coenzyme or distinguishing characteristic based on whether it corresponds to catalytic or stoichiometric coenzymes. Catalytic coenzymes Answer Bank Stoichiometric coenzymes lipoic acid FAD used once coenzyme A regenerated thiamine pyrophosphate (TPP) NAD+arrow_forward
- The oxidation of malate by NAD+ to form oxaloacetate is a highly endergonic reaction under standard conditions. AG +29 kJ mol¹ (+7 kcal mol-¹) Malate + NAD+ oxaloacetate + NADH + H+ The reaction proceeds readily under physiological conditions. = Why does the reaction proceed readily as written under physiological conditions? The NADH produced during glycolysis drives the reaction in the direction of malate oxidation. The steady-state concentrations of the products are low compared with those of the substrates. The reaction is pushed forward by the energetically favorable oxidation of fumarate to malate. Endergonic reactions such as this occur spontaneously without the input of free energy. Assuming an [NAD+ ]/[NADH] ratio of 8, a temperature of 25°C, and a pH of 7, what is the lowest [malate]/[oxaloacetate] ratio at which oxaloacetate can be formed from malate? [malate] [oxaloacetate]arrow_forwardCalculate and compare the AG values for the oxidation of succinate by NAD+ and FAD. Use the data given in the table to find the E of the NAD+: NADH and fumarate:succinate couples, and assume that E for the enzyme-bound FAD: FADH2 redox couple is nearly +0.05 V. Oxidant Reductant " E' (V) NAD+ NADH + H+ 2 -0.32 Fumarate Succinate AG°' for the oxidation of succinate by NAD+: AG°' for the oxidation of succinate by FAD: 2 -0.03 Why is FAD rather than NAD+ the electron acceptor in the reaction catalyzed by succinate dehydrogenase? The electron-transport chain can regenerate FAD, but not NAD+. FAD is an oxidant, whereas NAD+ is a reductant. The oxidation of succinate requires two NAD+ molecules but only one FAD molecule. The oxidation of succinate by NAD+ is not thermodynamically feasible. kJ mol-1 kJ mol-1arrow_forwardUse the cellular respiration interactive to help you complete the passage. 2,4-dinitrophenol (DNP) was a popular ingredient in diet pills in the 1930s before it was discovered that moderate doses of the compound cause exceptionally high body temperature and even death. Complete the passage detailing how DNP's mechanism of action explains why it causes both high body temperature and weight loss. 2,4-dinitrophenol (DNP) causes of returning to the mitochondrial matrix through to pass directly across the inner mitochondrial membrane instead proteins. Because of DNP's effect on the mitochondrion, less energy is captured in the form of energy is instead wasted as heat. and more protons electrons ATP NADH sugars cytochrome ATP synthase heatarrow_forward
- To answer this question, you may reference the Metabolic Map. Select the reactions of glycolysis in which ATP is produced. 1,3-Bisphosphoglycerate 3-phosphoglycerate Glyceraldehyde 3-phosphate 1,3-bisphosphoglycerate Fructose 6-phosphate fructose 1,6-bisphosphate Phosphoenolpyruvate pyruvate Glucose glucose 6-phosphate Suppose 17 glucose molecules enter glycolysis. Calculate the total number of inorganic phosphate (P) molecules required as well as the total number of pyruvate molecules produced. P required: pyruvate produced: molecules moleculesarrow_forwardSuppose a marathon runner depletes carbohydrate stores after a four-hour run. The runner's nutritionist suggests replenishing carbohydrate stores by eating carbohydrates. However, the runner is also concerned about weight loss and wants to know if fats can be directly converted into carbohydrates. How should the nutritionist respond to the runner? Yes, the glyoxylate cycle can be used to convert acetyl CoA into succinate, which can then be converted into carbohydrates. No, the two decarboxylation reactions of the citric acid cycle preclude the net conversion of acetyl CoA into carbohydrates. No, the citric acid cycle converts acetyl CoA into oxaloacetate, but there is no pathway to form glucose from oxaloacetate. Yes, pyruvate carboxylase can convert acetyl CoA into pyruvate, which can be used to form glucose through gluconeogenesis.arrow_forwardThe crossover technique can reveal the precise site of action of a respiratory-chain inhibitor. Britton Chance devised elegant spectroscopic methods for determining the proportions of the oxidized and reduced form of each carrier. This determination is feasible because the forms have distinctive absorption spectra, as illustrated in the graph for cytochrome c. Upon the addition of a new inhibitor to respiring mitochondria, the carriers between NADH and ubiquinol (QH2) become more reduced, and those between cytochrome c and O₂ become more oxidized. Where does your inhibitor act? Complex I Complex II Complex III Complex IV Absorbance coefficient (M-1 cm x 10-5) 10 1.0 0.5 400 Reduced Oxidized 500 Wavelength (nm) 600arrow_forward
- Why are the electrons carried by FADH2 not as energy rich as those carried by NADH? FADH2 carries fewer high-energy electrons than NADH. OFADH2 is less negatively charged than NADH. OFADH2 has a lower phosphoryl-transfer potential than NADH. FADH₂ has a lower reduction potential than NADH. What is the consequence of this difference? Electrons flow from NADH to FADH2 before they are transferred to O₂. Electron flow FADH₂ to O, results in the production of more ATP than does electron flow from NADH. Electron flow from FADH₂ to O, pumps fewer protons than does electron flow from NADH. Electron flow from FADH, to O, consumes more free energy than does electron flow from NADH. A simple equation relates the standard free-energy change, AG", to the change in reduction potential, AE. AG=-FAE Then represents the number of transferred electrons, and F is the Faraday constant with a value of 96.48 kJ mol¹ V-¹. Use the standard reduction potentials provided to determine the standard free energy…arrow_forwardMatch each enzyme with its description. catalyzes the formation of isocitrate synthesizes succinyl CoA generates malate generates ATP converts pyruvate into acetyl CoA converts pyruvate into oxaloacetate condenses oxaloacetate and acetyl CoA catalyzes the formation of oxaloacetate synthesizes fumarate catalyzes the formation of a-ketoglutarate Answer Bank succinate dehydrogenase a-ketoglutarate dehydrogenase aconitase fumarase citrate synthase malate dehydrogenase pyruvate carboxylase pyruvate dehydrogenase complex isocitrate dehydrogenase succinyl CoA synthetasearrow_forwardcoo ☐ CH2 coo Malonate Determine how the concentration of each citric acid cycle intermediate will change immediately after the addition of malonate. The concentration of citrate will The concentration of isocitrate will The concentration of α-ketoglutarate will The concentration of succinyl CoA will The concentration of succinate will The concentration of fumarate will The concentration of malate will The concentration of oxaloacetate will Why is malonate not a substrate for succinate dehydrogenase? Malonate lacks a thioester bond that has high transfer potential. Malonate has two carboxylic acid groups. Malonate is not large enough to bind to the enzyme. Malonate only has one methylene group.arrow_forward
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON





