
Interpretation:
The structure of an alcohol of given molecular formula C4H10O to be predicted using 13CNMR spectra.
Concept introduction:
The 13CNMR spectrum gives information on the different electronic environments of carbon. As like 1HNMR, the number of signals generated in 13CNMR are predicted by performing symmetry operations (rotation or reflection symmetry). Only chemical shift values are reported in the spectrum but not the multiplicity and integration values because the coupling between two neighboring 13C-13 C nuclei are weakly involved due to the low abundance of 13C isotopes of carbon atom.
To Identify:
The structure of an alcohol of given molecular formula C8H16O.
Broadband-decoupled 13CNMR spectrum:
The spectra provide information regarding the total number of carbon environments.
DEPT (Distortionless enhancement by polarization transfer):
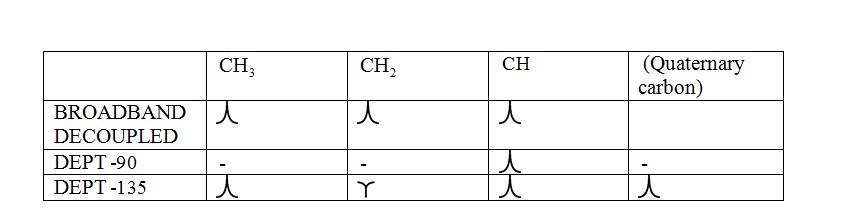
a) DEPT-90: The spectrum exhibits signal only from CH group and no signals from CH3, CH2, CH and quaternary carbon (carbon with no protons).
b) DEPT-135: The spectrum exhibits CH3 groups and CH groups as positive signals (pointing up); CH2 groups appear as negative signals (pointing down) and quaternary carbon does not appear.
The signals appear in each type of spectrum:
Trending nowThis is a popular solution!

Chapter 13 Solutions
OWLv2 with Student Solutions Manual eBook, 4 terms (24 months) Printed Access Card for McMurry's Organic Chemistry, 9th
- Using wedge-and-dash bonds, modify the bonds on the chiral carbon in the molecule below so the molecule has R stereochemical configuration. NH H Br X टेarrow_forwardProvide photos of models of the following molecules. (Include a key for identification of the atoms) 1,2-dichloropropane 2,3,3-trimethylhexane 2-bromo-3-methybutanearrow_forwardPlease draw the structure in the box that is consistent with all the spectral data and alphabetically label all of the equivalent protons in the structure (Ha, Hb, Hc....) in order to assign all the proton NMR peaks. The integrations are computer generated and approximate the number of equivalent protons. Molecular formula: C13H1802 14 13 12 11 10 11 (ppm) Structure with assigned H peaks 2.08 3.13arrow_forward
- A 0.10 M solution of acetic acid (CH3COOH, Ka = 1.8 x 10^-5) is titrated with a 0.0250 M solution of magnesium hydroxide (Mg(OH)2). If 10.0 mL of the acid solution is titrated with 10.0 mL of the base solution, what is the pH of the resulting solution?arrow_forwardFirefly luciferin exhibits three rings. Identify which of the rings are aromatic. Identify which lone pairs are involved in establishing aromaticity. The lone pairs are labeled A-D below.arrow_forwardA 0.10 M solution of acetic acid (CH3COOH, Ka = 1.8 x 10^-5) is titrated with a 0.0250 M solution of magnesium hydroxide (Mg(OH)2). If 10.0 mL of the acid solution is titrated with 10.0 mL of the base solution, what is the pH of the resulting solution?arrow_forward
- Given a complex reaction with rate equation v = k1[A] + k2[A]2, what is the overall reaction order?arrow_forwardPlease draw the structure in the box that is consistent with all the spectral data and alphabetically label all of the equivalent protons in the structure (Ha, Hb, Hc....) in order to assign all the proton NMR peaks. The integrations are computer generated and approximate the number of equivalent protons. Molecular formula: C13H1802 14 13 12 11 10 11 (ppm) Structure with assigned H peaks 2.08 3.13arrow_forwardCHEMICAL KINETICS. One of the approximation methods for solving the rate equation is the steady-state approximation method. Explain what it consists of.arrow_forward
- CHEMICAL KINETICS. One of the approximation methods for solving the rate equation is the limiting or determining step approximation method. Explain what it consists of.arrow_forwardCHEMICAL KINETICS. Indicate the approximation methods for solving the rate equation.arrow_forwardTRANSMITTANCE เบบ Please identify the one structure below that is consistent with the 'H NMR and IR spectra shown and draw its complete structure in the box below with the protons alphabetically labeled as shown in the NMR spectrum and label the IR bands, including sp³C-H and sp2C-H stretch, indicated by the arrows. D 4000 OH LOH H₂C CH3 OH H₂C OCH3 CH3 OH 3000 2000 1500 HAVENUMBERI-11 1000 LOCH3 Draw your structure below and label its equivalent protons according to the peak labeling that is used in the NMR spectrum in order to assign the peaks. Integrals indicate number of equivalent protons. Splitting patterns are: s=singlet, d=doublet, m-multiplet 8 3Hb s m 1Hd s 3Hf m 2Hcd 2Had 1He 鄙视 m 7 7 6 5 4 3 22 500 T 1 0arrow_forward
 Introduction to General, Organic and BiochemistryChemistryISBN:9781285869759Author:Frederick A. Bettelheim, William H. Brown, Mary K. Campbell, Shawn O. Farrell, Omar TorresPublisher:Cengage Learning
Introduction to General, Organic and BiochemistryChemistryISBN:9781285869759Author:Frederick A. Bettelheim, William H. Brown, Mary K. Campbell, Shawn O. Farrell, Omar TorresPublisher:Cengage Learning
