
Concept explainers
If a restriction enzyme cuts between the G and the A whenever it encounters the sequence GAATTC, how many fragments will be produced when the enzyme is digested with DNA with the following sequence? TGAGAATTCAACTGAATTCAAATTCGAATTCTTAGC
- a. Two
- b. Three
- c. Four
- d. Five
Introduction:
Restriction enzymes are the enzymes that digest the higher molecular weight DNA in to smaller fragments. They are also known as restriction endonucleases.
Answer to Problem 1MCQ
Correct answer:
There are three sequences of GAATTC present on the DNA fragment and the fragments formed after the cleavage of the DNA are four in numbers. Therefore, option (c) is correct.
Explanation of Solution
Reason for correct statement:
Restriction endonucleases digest a double stranded DNA at specific sites. These specific sites are also called recognition site that present as palindrome. If the restriction enzyme given cuts between the G and A whenever it encounters the sequence GAATTC on the given DNA with the following sequence TGAGAATTCTGAATTCAAATTCGAATTCTTAGC, the cleaving of the DNA will be given as follows:
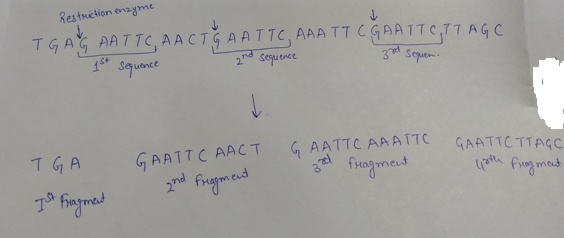
As shown, after the cleavage of the DNA by the help of the restriction enzyme, four fragments will be formed.
Option (c) is given as “Four”.
As, “the restriction enzyme recognizes three sequence on the given DNA, so it will make three cuts, that results in the production of the four DNA fragments,” is the right answer.
Hence, option (c) is correct.
Reasons for the incorrect statements:
Option (a), is given as “Two”.
The fragments of the DNA formed will be four after the action of the restriction enzyme. Hence, it is a wrong answer.
Option (b), is given as “Three”.
The DNA fragment formed after the cleavage will be four in numbers. Hence, it is a wrong answer.
Option (d), is given as “Five”.
For DNA fragments will be formed after the cutting action of the restriction enzyme at the given recognition site. Hence, it is a wrong answer.
Hence, options (a), (b), and (d), are incorrect.
Restriction enzymes are also known as molecular scissors that could cut double stranded DNA molecules at specific sites. It is an important tool that is used in the manipulation of DNA.
Want to see more full solutions like this?
Chapter 11 Solutions
Biology: Concepts and Investigations
Additional Science Textbook Solutions
Physics for Scientists and Engineers
Introductory Chemistry (6th Edition)
Laboratory Manual For Human Anatomy & Physiology
Microbiology Fundamentals: A Clinical Approach
Human Physiology: An Integrated Approach (8th Edition)
- When beta-lactamase was isolated from Staphylcoccus aureus and treated with a phosphorylating agent, only the active site, serine was phosphorylated. Additionally, the serine was found to constitute 0.35% (by weight) of this beta-lactamase enzyme. Using this, calculate the molecular weight of this enzyme and estimate the number of amino acids present in the polypeptide.arrow_forwardBased on your results from the Mannitol Salt Agar (MSA) media, which of your bacteria were mannitol fermenters and which were not mannitol fermenters?arrow_forwardhelp tutor pleasearrow_forward
- Q8. A researcher wants to study the effectiveness of a pill intended to reduce stomach heartburn in pregnant women. The researcher chooses randomly 400 women to participate in this experiment for 9 months of their pregnancy period. They all need to have the same diet. The researcher designs two groups of 200 participants: One group take the real medication intended to reduce heartburn, while the other group take placebo medication. In this study what are: Independent variable: Dependent variable: Control variable: Experimental group: " Control group: If the participants do not know who is consuming the real pills and who is consuming the sugar pills. This study is It happens that 40% of the participants do not find the treatment helpful and drop out after 6 months. The researcher throws out the data from subjects that drop out. What type of bias is there in this study? If the company who makes the medication funds this research, what type of bias might exist in this research work?arrow_forwardHow do I determine the inhertiance pattern from the pedigree diagram?arrow_forwardits an open book assignemntarrow_forward
- Describe two different gene regulation mechanisms involving methylationarrow_forwardWhat is behavioral adaptarrow_forward22. Which of the following mutant proteins is expected to have a dominant negative effect when over- expressed in normal cells? a. mutant PI3-kinase that lacks the SH2 domain but retains the kinase function b. mutant Grb2 protein that cannot bind to RTK c. mutant RTK that lacks the extracellular domain d. mutant PDK that has the PH domain but lost the kinase function e. all of the abovearrow_forward
 Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning





