
Genetics: From Genes To Genomes (6th International Edition)
6th Edition
ISBN: 9781260041217
Author: Leland Hartwell Dr., ? Michael L. Goldberg Professor Dr., ? Janice Fischer, ? Leroy Hood Dr.
Publisher: Mcgraw-Hill
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 10, Problem 3P
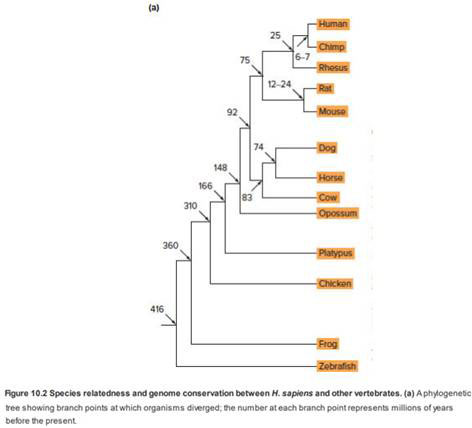
Figure 10.2a has numbers indicating the approximate number of millions of years ago that species on separate branches of the tree last shared a common ancestor.
| a. | About how many millions of years ago did humans last share a common ancestor with chimpanzees, mice, dogs, chickens, and frogs? These estimates for evolutionary events were obtained in part by comparing the genomic sequences of various current-day species. The basic supposition behind these estimates is that of a molecular clock: Differences in particular types of genomic sequences accumulate at a relatively linear rate during evolutionary time. Consider the three following kinds of |
| b. | . . . represent the slowest-ticking clock? (That is, which type of mutation would accumulate the least rapidly in genomes? Hint: See Fig. 10.3.) |
| c. | . . . you most likely use to estimate the divergence times of species that last shared a common ancestor more than 400 million years ago? |
| d. | . . . be most likely to vary in the rate at which they would accumulate in different genes? |
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Which microbe does not live part of its lifecycle outside humans?
A. Toxoplasma gondii
B. Cytomegalovirus
C. Francisella tularensis
D. Plasmodium falciparum
explain your answer thoroughly.
Select all of the following that the ablation (knockout) or ectopoic expression (gain of function) of Hox can contribute to.
Another set of wings in the fruit fly, duplication of fingernails, ectopic ears in mice, excess feathers in duck/quail chimeras, and homeosis of segment 2 to jaw in Hox2a mutants
Select all of the following that changes in the MC1R gene can lead to:
Changes in spots/stripes in lizards, changes in coat coloration in mice, ectopic ear formation in Siberian hamsters, and red hair in humans
Chapter 10 Solutions
Genetics: From Genes To Genomes (6th International Edition)
Ch. 10 - Prob. 1PCh. 10 - List three independent techniques you could use to...Ch. 10 - Figure 10.2a has numbers indicating the...Ch. 10 - Which of the enzymes from the following list would...Ch. 10 - Prob. 5PCh. 10 - a. What sequence information about a gene is...Ch. 10 - Why do geneticists studying eukaryotic organisms...Ch. 10 - Consider three different kinds of human libraries:...Ch. 10 - The human genome has been sequenced, but we still...Ch. 10 - This problem investigates issues encountered in...
Ch. 10 - For the sake of simplicity, Fig. 10.4 omitted one...Ch. 10 - Give two different reasons for the much higher...Ch. 10 - Using a cDNA library, you isolated two different...Ch. 10 - The figure that follows shows part of a modified...Ch. 10 - In Problem 14, cDNAs F and G could not be found in...Ch. 10 - Fig. 10.10 presents a model for exon shuffling in...Ch. 10 - An interesting phenomenon found in vertebrate DNA...Ch. 10 - a. If you found a zinc-finger domain which...Ch. 10 - Prob. 19PCh. 10 - In the human immune system, so-called B cells can...Ch. 10 - Chimpanzees have a set of hemoglobin genes very...Ch. 10 - Complete genome sequences indicate that the human...Ch. 10 - On your computers browser, view the page accessed...Ch. 10 - Prob. 24PCh. 10 - Prob. 25PCh. 10 - Certain individuals with mild forms of...Ch. 10 - The 1 and 2 genes in humans are identical in their...Ch. 10 - Prob. 28P
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Pleiotropic genes are genes that (blank) Cause a swapping of organs/structures, are the result of duplicated sets of chromosomes, never produce protein products, and have more than one purpose/functionarrow_forwardA loss of function mutation in Pitx1 enhancers can cause (blank) Removal of Pitx1 exons and growth of ectopic hindlimbs, growth of extra ectopic forelimbs, loss of forelimb specification and development, and loss of hindlimb specification and developmentarrow_forwardHox1a most likely contributes to (blank) patterning in the developing embryo? Ventral, posterior, limb or anteriorarrow_forward
- Select all of the following that can help establish Hox gene expression boundaries (things that affect Hox and not things that Hox affects). Retinoic acid, anterior/posterior axis, fibroblast growth factors, vagal neural crest, and enhancersarrow_forwardEctopic expression of Hox often results in (blank) phenotypes. (Blank) transformations are characterized by the replacement of one body part/structure with another. Hoxeotic, homealoneotic, joexotic, or homeoticarrow_forwardWhat's the difference when drawing omega-6 and omega-3?arrow_forward
- . Consider a base substitution mutation that occurred in a DNA sequence that resulted in a change in the encoded protein from the amino acid glutamic acid to aspartic acid. Normally the glutamic acid amino acid is located on the outside of the soluble protein but not near an active site. O-H¨ A. What type of mutation occurred? O-H B. What 2 types of chemical bonds are found in the R-groups of each amino acid? The R groups are shaded. CH2 CH2 CH2 H2N-C-COOH H2N-C-COOH 1 H Glutamic acid H Aspartic acid C. What 2 types of bonds could each R-group of each of these amino acids form with other molecules? D. Consider the chemical properties of the two amino acids and the location of the amino acid in the protein. Explain what effect this mutation will have on this protein's function and why.arrow_forwardengineered constructs that consist of hollow fibers are acting as synthetic capillaries, around which cells have been loaded. The cellular space around a single fiber can be modeled as if it were a Krogh tissue cylinder. Each fiber has an outside “capillary” radius of 100 µm and the “tissue” radius can be taken as 200 µm. The following values apply to the device:R0 = 20 µM/secaO2 = 1.35 µM/mmHgDO2,T = 1.67 x 10-5 cm2/secPO2,m = 4 x 10-3 cm/secInstead of blood inside the fibers, the oxygen transport and tissue consumption are being investigated by usingan aqueous solution saturated with pure oxygen. As a result, there is no mass transfer resistance in the synthetic“capillary”, only that due to the membrane itself. Rather than accounting for pO2 variations along the length ofthe fiber, use an average value in the “capillary” of 130 mmHg.Is the tissue fully oxygenated?arrow_forwardMolecular Biology Please help with question. thank you You are studying the expression of the lac operon. You have isolated mutants as described below. In the presence of glucose, explain/describe what would happen, for each mutant, to the expression of the lac operon when you add lactose AND what would happen when the bacteria has used up all of the lactose (if the mutant is able to use lactose).5. Mutations in the lac operator that strengthen the binding of the lac repressor 200 fold 6. Mutations in the promoter that prevent binding of RNA polymerase 7. Mutations in CRP/CAP protein that prevent binding of cAMP8. Mutations in sigma factor that prevent binding of sigma to core RNA polymerasearrow_forward
- Molecular Biology Please help and there is an attached image. Thank you. A bacteria has a gene whose protein/enzyme product is involved with the synthesis of a lipid necessary for the synthesis of the cell membrane. Expression of this gene requires the binding of a protein (called ACT) to a control sequence (called INC) next to the promoter. A. Is the expression/regulation of this gene an example of induction or repression?Please explain:B. Is this expression/regulation an example of positive or negative control?C. When the lipid is supplied in the media, the expression of the enzyme is turned off.Describe one likely mechanism for how this “turn off” is accomplished.arrow_forwardMolecular Biology Please help. Thank you. Discuss/define the following:(a) poly A polymerase (b) trans-splicing (c) operonarrow_forwardMolecular Biology Please help with question. Thank you in advance. Discuss, compare and contrast the structure of promoters inprokaryotes and eukaryotes.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Biology (MindTap Course List)
Biology
ISBN:9781337392938
Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. Berg
Publisher:Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning


Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning
Discovering the tree of life | California Academy of Sciences; Author: California Academy of Sciences;https://www.youtube.com/watch?v=AjvLQJ6PIiU;License: Standard Youtube License