
Concept explainers
DNA is not the only molecule that is used to test for evolutionary relationships among organisms. Proteins can also be used, and the sequences of their building blocks (called amino acids) can be compared in much the same way that DNA sequences are compared. Cytochrome c is a protein found in nearly all living organisms; it functions in the transformation of energy within cells. The percentage difference in amino acid sequence between humans and other organisms can be summarized as follows:
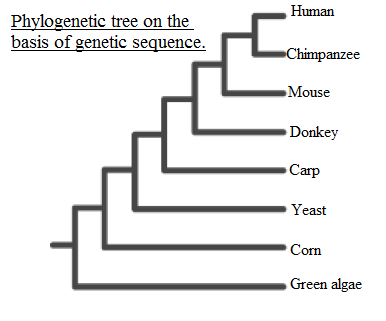
Draw the evolutionary tree implied by these data that illustrates the relationship between humans and the other organisms listed.
Want to see the full answer?
Check out a sample textbook solution
Chapter 10 Solutions
EBK BIOLOGY
Additional Science Textbook Solutions
Biological Science (6th Edition)
Genetics: From Genes to Genomes
Microbiology Fundamentals: A Clinical Approach
Human Physiology: An Integrated Approach (8th Edition)
Campbell Essential Biology (7th Edition)
- can you help? I think its B but not surearrow_forwardSkip to main content close Homework Help is Here – Start Your Trial Now! arrow_forward search SEARCH ASK Human Anatomy & Physiology (11th Edition)BUY Human Anatomy & Physiology (11th Edition) 11th Edition ISBN: 9780134580999 Author: Elaine N. Marieb, Katja N. Hoehn Publisher: PEARSON 1 The Human Body: An Orientation expand_moreChapter 1 : The Human Body: An Orientation Chapter Questions expand_moreSection: Chapter Questions Problem 1RQ: The correct sequence of levels forming the structural hierarchy is A. (a) organ, organ system,... format_list_bulletedProblem 1RQ: The correct sequence of levels forming the structural hierarchy is A. (a) organ, organ system,... See similar textbooks Bartleby Related Questions Icon Related questions Bartleby Expand Icon bartleby Concept explainers bartleby Question Draw a replication bubble with two replication forks.blue lines are DNA single strands and red lines are RNA single strands.indicate all 3' and 5’ ends on all DNA single…arrow_forwardProvide an answerarrow_forward
- Question 4 1 pts Which of the following would be most helpful for demonstrating alternative splicing for a new organism? ○ its proteome and its transcriptome only its transcriptome only its genome its proteome and its genomearrow_forwardIf the metabolic scenario stated with 100 mM of a sucrose solution, how much ATP would be made then during fermentation?arrow_forwardWhat is agricuarrow_forward
- When using the concept of "a calorie in is equal to a calorie out" how important is the quality of the calories?arrow_forwardWhat did the Cre-lox system used in the Kikuchi et al. 2010 heart regeneration experiment allow researchers to investigate? What was the purpose of the cmlc2 promoter? What is CreER and why was it used in this experiment? If constitutively active Cre was driven by the cmlc2 promoter, rather than an inducible CreER system, what color would you expect new cardiomyocytes in the regenerated area to be no matter what? Why?arrow_forwardWhat kind of organ size regulation is occurring when you graft multiple organs into a mouse and the graft weight stays the same?arrow_forward
- What is the concept "calories consumed must equal calories burned" in regrads to nutrition?arrow_forwardYou intend to insert patched dominant negative DNA into the left half of the neural tube of a chick. 1) Which side of the neural tube would you put the positive electrode to ensure that the DNA ends up on the left side? 2) What would be the internal (within the embryo) control for this experiment? 3) How can you be sure that the electroporation method itself is not impacting the embryo? 4) What would you do to ensure that the electroporation is working? How can you tell?arrow_forwardDescribe a method to document the diffusion path and gradient of Sonic Hedgehog through the chicken embryo. If modifying the protein, what is one thing you have to consider in regards to maintaining the protein’s function?arrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning
Biology (MindTap Course List)BiologyISBN:9781337392938Author:Eldra Solomon, Charles Martin, Diana W. Martin, Linda R. BergPublisher:Cengage Learning Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College





