
EBK BIOLOGY
6th Edition
ISBN: 9780134819075
Author: Maier
Publisher: PEARSON CUSTOM PUB.(CONSIGNMENT)
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 9, Problem 9LTB
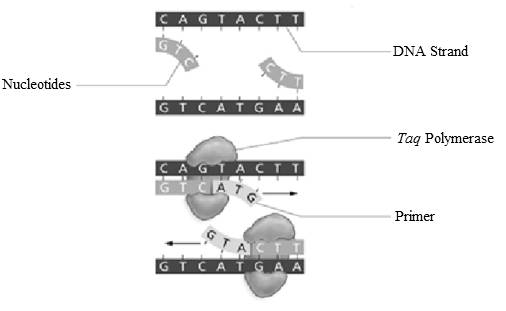
Add labels to the figure that follows, which illustrates the components in the PCR reaction.
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Look at each PCR component listed below. For each one, determine which steps(s) of the PCR reaction (denaturation, annealing or extension) would be directly affect if that component were missing.
Taq polymerase:
Oligonucleotide primers:
DNA template:
Deoxynucleotides (A, T, G and C):
Imagine that you correctly prepare your PCR reaction mixture, but there is something wrong with the thermal cycler. Describe what would happen if:
The thermal cycler was stuck on 940C:
The thermal cycler cycled between 600C and 720C, but never reached 940C
The thermal cycler cycled between 940C and 720C, but never reached 600
This is what the LAB FLOW would look like:
CULTURE BACTERIA
EXTRACT RNA
RANDOM HEXAMER REVERSE TRANSCRIPTION OF RNA
PCR OF lacZ GENE
ELECTROPHORESIS
If you wanted to prepare only one PCR reaction, how much of each reagent would you add to the PCR tube?
What would be the final primer concentration if 0.5 μl of 10 μM primers were added to a PCR reaction with a final volume of 20 μl?
Explain why a positive control and negative control are included in PCR experiments.
Explain the three steps involved in each cycle of polymerase chain reaction.Why is loading dye added to the DNA sample for gel electrophoresis?
Explain the function of the following components in a PCR reaction:− Primer, dNTP, MgCl, Taq polymerase, buffer.
Chapter 9 Solutions
EBK BIOLOGY
Ch. 9 - Is a round yellow pea seed (genotype Rr Y y) an...Ch. 9 - What factors cause quantitative variation in a...Ch. 9 - The DNA profile below is from a mother, a father,...Ch. 9 - Prob. 4LTBCh. 9 - Prob. 5LTBCh. 9 - Prob. 6LTBCh. 9 - Prob. 7LTBCh. 9 - Prob. 8LTBCh. 9 - Add labels to the figure that follows, which...Ch. 9 - Prob. 10LTB
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Which of the following best describes the process of DNA sequencing? a. DNA is separated on a gel, and the different bands are labeled with fluorescent nucleotides and scanned with a laser. b. A laser is used to fluorescently label the nucleotides present within the DNA, the DNA is run on a gel, and then the DNA is broken into fragments. c. Nucleotides are scanned with a laser and incorporated into the DNA that has been separated on a gel, and then the DNA is amplified with PCR. d. Fragments of DNA are produced in a reaction that labels them with any of four different fluorescent dyes, and the fragments then are run on a gel and scanned with a laser. e. DNA is broken down into its constituent nucleotides, and the nucleotides are then run on a gel and purified with a laser.arrow_forwardYou want to set up 50 µl total volume of a PCR reaction. You have a microfuge tube of Taq polymerase (5 units/ul), and each PCR reaction requires a final of 10 units of Taq polymerase. You have a microfuge tube of Taq Buffer (5X), and each PCR reaction requires a final of 1X Taq Buffer. How much of Taq polymerase and Taq buffer would you add? Select all that apply 10 ul of Taq buffer 2 ul of Taq polymerase 5 ul of Taq buffer 5 ul of Taq polymerasearrow_forwardBelow are several problems frequently faced by researchers when running the PCR. Give one (1) solution to each problem stated. 1. No PCR product 2. Multiple bands appeared after gel electrophoresis (you are amplifying just 1 gene) 3. Bright thick bands at the end of the agarose gel after electrophoresis 4. Bands appeared at the negative controlarrow_forward
- After running a qPCR experiment, we will have graphs showing the amount of fluorescence detected by the digital camera compared to the number of PCR cycles run. Suppose you see the following graph output by the qPCR machine: Relative Fluoresence 3.0 2.5 2.0 1.5 1.0 0.5 0.0 0 10 20 30 40 50 Cycles Which curve (blue, red, or green) represents a sample with the smallest amount of mRNA present? Why? Be sure to discuss Ct values in your answer.arrow_forwardYou want to set up 50 µl total volume of a PCR reaction. You have a microfuge tube of Forward primer (25 µM), and each PCR reaction requires a final of 1 uM of primer. You have a microfuge tube of Reverse primer (25 µM), and each PCR reaction requires a final of 1 µM of primer. How much of the primers would you add? Select all that apply 2 ul of Reverse primer 2 ul of Forward primer 5 ul of Forward primer 5 ul of Reverse primerarrow_forwardThe process of PCR essentially revolves around three phases. Briefly describe these phases and the events that occur in them. Take note the temperature on which these phases take place.arrow_forward
- There are many types of PCR techniques available nowadays. Explain the concept of nested PCR and real-time PCR.arrow_forwardExplain the importance of different temperatures in PCR. Tm of forward primer= 62 °C Tm of reverse primer= 59 °C Gene size= 3 Kbarrow_forwardExplain the importance of different temperatures in PCR.Tm of forward primer= 62 °CTm of reverse primer= 59 °CGene size= 3 Kbarrow_forward
- The gel below shows results for the bitter tasting SNP analysis from a CH306 class from a few years ago. Analyze the results and annotate the gel to indicate the bitter tasting ability and homozygous/heterozygous status of each of the individuals represented on the gel. There are three lanes of markers, and the first non-maker lane on the left of the gel is an uncut control PCR product (i.e. there are a total of 10 individuals on the gel starting in lane 3).arrow_forwardThe amount of each component for a PCR reaction is critical. The final concentration of each primer should be 0.2 uM, and the final concentration of dNTPs is 0.2 mM. If the final concentrations of these components are significantly off, your PCR reactions will not work. The stock concentration of primers is usually 10 uM and the stock concentration of dNTPs is usually 10 mM. PCR buffer is supplied as 10X and should be 1X in the final reaction volume. The amount of Taq DNA polymerase necessary will depend on the manufacturer. For our purposes, Taq polymerase is supplied at a concentration of 20 Units/ ul. Each PCR reaction should include 5 units of Taq polymerase. 1) Calculate the necessary volume to set up a single PCR reaction given the following: Assume you will use 2 ul of template DNA (e.g. isolated from a patient). You have the following primer stocks GAPDH Forward Primer, 10 uM stock GAPDH Reverse Primer, 10 uM stock SARS-CoV-2 Forward Primer, 10 uM stock SARS-CoV-2 Reverse…arrow_forwardAll things considered, the most important factor with respect to successful PCR is Multiple Choice primer design annealing temperature extension time quantity of templatearrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning
Molecular Techniques: Basic Concepts; Author: Dr. A's Clinical Lab Videos;https://www.youtube.com/watch?v=7HFHZy8h6z0;License: Standard Youtube License