
Concept explainers
To gauge the fidelity of DNA synthesis, Arthur Kornberg and colleagues devised a technique called nearest-neighbor analysis, which determines the frequency with which any two bases occur adjacent to each other along the polynucleotide chain (J. Biol. Chem. 236: 864–875). This test relies on the enzyme spleen phosphodiesterase (see the previous problem). As we saw in Figure 11-8, DNA is synthesized by
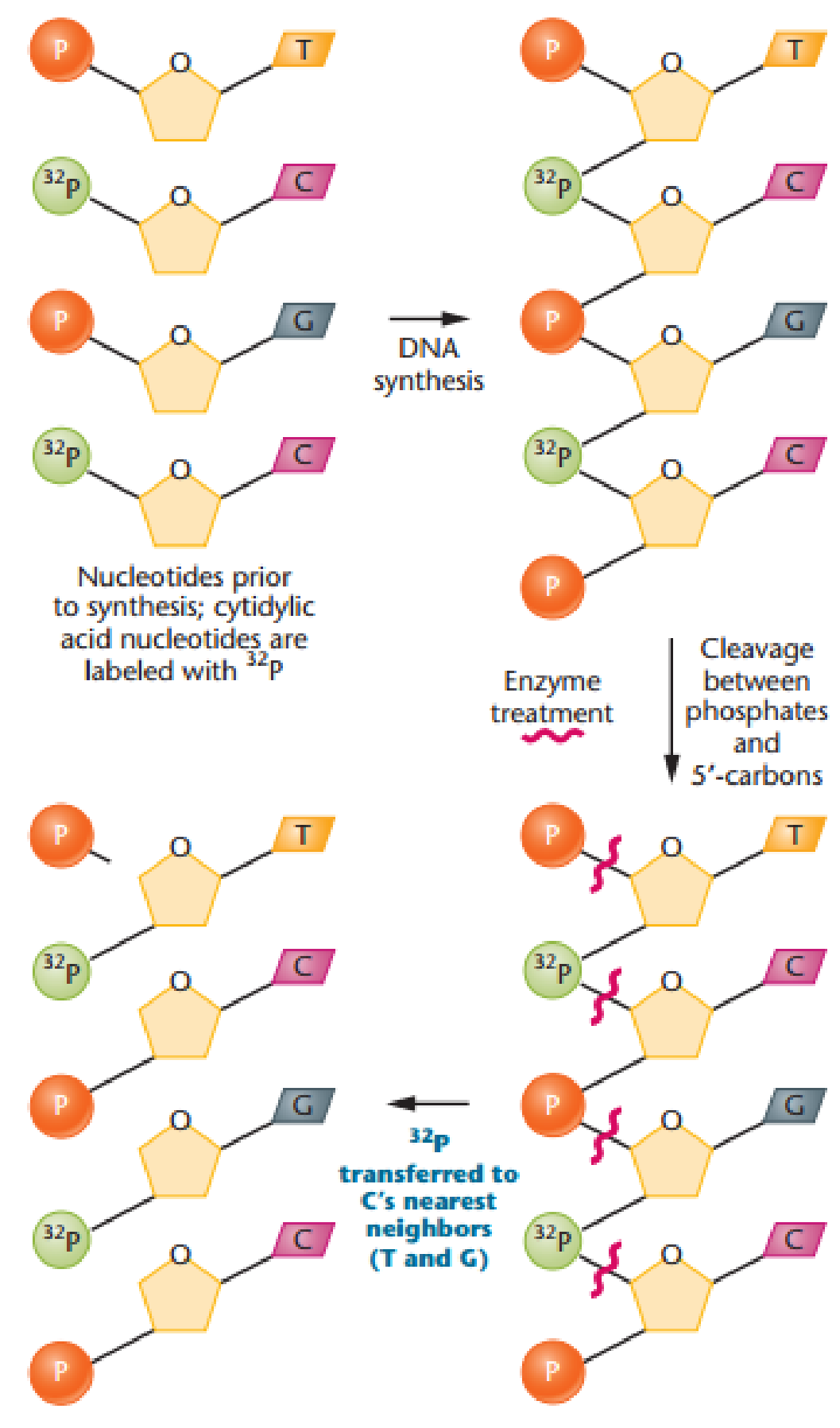
Following four separate experiments, in each of which a different one of the four nucleotide types is radioactive, the frequency of all 16 possible nearest neighbors can be calculated. When Romberg applied the nearest-neighbor frequency test to the DNA template and resultant product from a variety of experiments, he found general agreement between the nearest-neighbor frequencies of the two.
Analysis of nearest-neighbor data led Kornberg to conclude that the two strands of the double helix are in opposite polarity to one another. Demonstrate this approach by determining the outcome of such an analysis if the strands of DNA shown here are (a) antiparallel versus (b) parallel:

Trending nowThis is a popular solution!

Chapter 11 Solutions
EBK CONCEPTS OF GENETICS
- The BamH1 enzyme comes at a concentration of 100,000 U/ml. You are asked to digest 20 ug of DNA with this enzyme. Determine: a) How many units will you need? b) How will you dispense them?arrow_forwardUse a drawing to illustrate the principle of DNA gel electrophoresis. (2 marks)-+arrow_forwardWhy was it necessary to mash the strawberries extensively with your hands? (hint: you wouldn’t have to do this step with an animal cells). Why do we treat the cells with soap when conducting DNA extraction? And why do we add salt when doing the DNA extraction?arrow_forward
- At higher amounts of protein, the Bradford assay is not linear. Consider the plot to the right: what is the maximum amount of protein a sample could contain and still fall within a standard curve? Briefly explain your reasoning.arrow_forwardWhat entropic factor destabilizes helical DNA at high temperature?arrow_forwardWhat additive in the agarose gel solution allows the DNA to fluoresce (glow) under UV light? How does this work?arrow_forward
- One micromole of 48-nucleotide(nt) DNA was synthesized via solid phase synthesis. This sample contains salts and DNA strands in different lengths (32-48 nt). Please propose a procedure and instruments to purify and confirm only 48-nt DNA from the mixture?arrow_forwardA recombinant protein corresponding to 52 repeats of the peptide sequence YSPTSPS was covalently linked to agarose beads to make an affinity column. The beads were incubated with a nuclear extract and the protein:beads became phosphorylated. More nuclear extract was run over the column repeatedly and then the beads were washed extensively. The bound proteins were eluted from the column and when they were separated on a SDS-PAGE gel many bands appeared. What cellular process(es) might these proteins be involved in? Options: splicing polyadenylation DNA replication a and b only a, b and carrow_forwardWhat two enthalpic factors stabilize DNA in double-helical form at low temperature?arrow_forward
- What is meant by the term genetic testing? How do testing at theprotein level and testing at the DNA level differ? Describe fivedifferent techniques used in genetic testing.arrow_forwardARE THEY TRUE OR FALSE? a) In a caesium chloride density gradient centrifugation method performed in the presence of ethidium bromide, supercoiled plasmid DNA binds more ethidium bromide than linear DNA. B)Tth DNA polymerase shows DNA-dependent DNA polymerase activity as well as RNA-dependent DNA polymerase activity. C)Magnesium ions stimulate polymerase activity. Therefore, it is better to use the highest concentration of magnesium in PCR amplification. D)When lambda bacteriophage infects E. coli cell lysis never occurs, and the infected bacterium can continue to grow and divide. E)The copy number refers to the number of molecules of an individual plasmid that are normally found in a single bacterial cell. F)RNase H selectively hydrolyzes phosphodiester bonds of RNA molecules in RNA:DNA duplexes.arrow_forwardNote that the table provided shows a ligation using a molar ratio of 1:3 vector to insert. Write out the complete recipe for a 5:1 insert: vector ligation reaction, including the volumes of insert and vector you calculated above, and the volumes required for a 20 uL reaction: Ligase buffer Nuclease-free water T4 DNA ligasearrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
